遗传 ›› 2025, Vol. 47 ›› Issue (2): 172-182.doi: 10.16288/j.yczz.24-148
收稿日期:2024-05-23
修回日期:2024-08-17
出版日期:2025-02-20
发布日期:2024-08-19
通讯作者:
张力,博士,研究员,研究方向:基因组学。E-mail: zhangli@cibr.ac.cn基金资助:Received:2024-05-23
Revised:2024-08-17
Published:2025-02-20
Online:2024-08-19
Supported by:摘要:
在生命演化过程中,突变随机产生并被选择固定。同时物种逐步形成,产生各种生命形式。在传统演化理论体系中,突变被默认为遗传突变,体细胞突变仅在肿瘤、免疫和衰老等特定场景使用。选择在生命系统的多个层面发挥作用,包括基因、细胞、组织器官、个体、群体、物种,乃至生态系统。现代生命科学主流研究观点将遗传突变表述为基因型,将细胞类型及其他层面特征表述为表型,并发现表型由基因型和环境因素共同决定。目前,尚不清楚基因型和环境因素在细胞层面的作用机制,以及新细胞类型的产生和固定机制。本文从基因演化研究出发,依托现有演化理论体系,初步论述了细胞类型演化的理论定位和潜在的研究方向。
本文勘误:见 遗传, 2025, Vol. 47 (3): 341.
张力, 李川昀. 从基因演化到细胞类型演化的理论思考[J]. 遗传, 2025, 47(2): 172-182.
Li Zhang, Chuanyun Li. Theoretical thinking from gene evolution to cell type evolution[J]. Hereditas(Beijing), 2025, 47(2): 172-182.
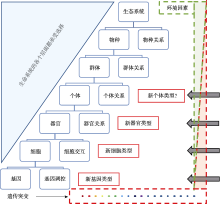
图1
生命演化的概念图 生命组织形式在演化过程中逐步变得复杂。复杂性增长呈现为二元树状结构。左外侧节点为每个层次的基本功能单位,不同基本功能单位的特定组合形成上一层次的基本功能单位。最底层面,基因被遗传突变直接影响,而基因调控被遗传突变和环境因素共同影响。特定基因的特定组合构成上一层次的细胞,这意味着细胞同时受遗传突变和环境因素影响。细胞间交互会被遗传突变和环境因素影响。特定细胞的特定组合构成上一层次的器官。以此类推,越高层次受到环境因素影响越大,相对遗传因素的影响占比就会减少。例如,同卵双胞胎在不同国家长大,在基因、细胞、器官层次的差异相对小,但是在个体认知方面差异巨大。也就是说,基于遗传信息对基因、细胞和器官的定义是相对稳定的,环境因素在个体层次开始主导。"

| [1] | Lewontin RC. The units of selection. Annual Review of Ecology and Systematics, 1970, 1: 1-18. |
| [2] | Wray NR, Visscher PM. Estimating trait heritability. Nat Educ, 2008, 1(1): 29. |
| [3] | Griffith F. The significance of pneumococcal types. J Hyg (Lond), 1928, 27(2): 113-159. |
| [4] | Watson JD, Crick FH. Molecular structure of nucleic acids: a structure for deoxyribose nucleic acid. Am J Psychiatry, 2003, 160(4): 623-624. |
| [5] | Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigó R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Deslattes Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X. The sequence of the human genome. Science, 2001, 291(5507): 1304-1351. |
| [6] | Eddy SR. The C-value paradox, junk DNA and ENCODE. Curr Biol, 2012, 22(21): R898-R899. |
| [7] |
Carroll SB. Endless forms: the evolution of gene regulation and morphological diversity. Cell, 2000, 101(6): 577-580.
pmid: 10892643 |
| [8] |
Chen SD, Krinsky BH, Long MY. New genes as drivers of phenotypic evolution. Nat Rev Genet, 2013, 14(9): 645-660.
doi: 10.1038/nrg3521 pmid: 23949544 |
| [9] | Chen SD, Zhang YE, Long MY. New genes in Drosophila quickly become essential. Science, 2010, 330(6011): 1682-1685. |
| [10] | Darwin CR. On the Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life. London: John Murray, 1859. |
| [11] |
Jacob F. Evolution and tinkering. Science, 1977, 196(4295): 1161-1166.
doi: 10.1126/science.860134 pmid: 860134 |
| [12] | Ohno S. Evolution by Gene Duplication. New York: Springer Berlin, Heidelberg, 1970. |
| [13] |
Wang W, Zhang J, Alvarez C, Llopart A, Long M. The origin of the Jingwei gene and the complex modular structure of its parental gene, yellow emperor, in Drosophila melanogaster. Mol Biol Evol, 2000, 17(9): 1294-1301.
pmid: 10958846 |
| [14] |
VanKuren NW, Long MY. Gene duplicates resolving sexual conflict rapidly evolved essential gametogenesis functions. Nat Ecol Evol, 2018, 2(4): 705-712.
doi: 10.1038/s41559-018-0471-0 pmid: 29459709 |
| [15] |
Zhou Q, Zhang GJ, Zhang Y, Xu SY, Zhao RP, Zhan ZB, Li X, Ding Y, Yang S, Wang W. On the origin of new genes in Drosophila. Genome Res, 2008, 18(9): 1446-1455.
doi: 10.1101/gr.076588.108 pmid: 18550802 |
| [16] |
Zhang YE, Vibranovski MD, Krinsky BH, Long MY. Age-dependent chromosomal distribution of male-biased genes in Drosophila. Genome Res, 2010, 20(11): 1526-1533.
doi: 10.1101/gr.107334.110 pmid: 20798392 |
| [17] | Zhang YE, Landback P, Vibranovski MD, Long MY. Accelerated recruitment of new brain development genes into the human genome. PLoS Biol, 2011, 9(10): e1001179. |
| [18] |
Domazet-Loso T, Brajković J, Tautz D. A phylostratigraphy approach to uncover the genomic history of major adaptations in metazoan lineages. Trends Genet, 2007, 23(11): 533-539.
pmid: 18029048 |
| [19] |
Long MY, VanKuren NW, Chen SD, Vibranovski MD. New gene evolution: little did we know. Annu Rev Genet, 2013, 47: 307-333.
doi: 10.1146/annurev-genet-111212-133301 pmid: 24050177 |
| [20] |
Tautz D, Domazet-Lošo T. The evolutionary origin of orphan genes. Nat Rev Genet, 2011, 12(10): 692-702.
doi: 10.1038/nrg3053 pmid: 21878963 |
| [21] |
Schlötterer C. Genes from scratch--the evolutionary fate of de novo genes. Trends Genet, 2015, 31(4) 215-219.
doi: 10.1016/j.tig.2015.02.007 pmid: 25773713 |
| [22] | Carvunis AR, Rolland T, Wapinski I, Calderwood MA, Yildirim MA, Simonis N, Charloteaux B, Hidalgo CA, Barbette J, Santhanam B, Brar GA, Weissman JS, Regev A, Thierry-Mieg N, Cusick ME, Vidal M. Proto-genes and de novo gene birth. Nature, 2012, 487(7407): 370-374. |
| [23] |
Zhao L, Saelao P, Jones CD, Begun DJ. Origin and spread of de novo genes in Drosophila melanogaster populations. Science, 2014, 343(6172): 769-772.
doi: 10.1126/science.1248286 pmid: 24457212 |
| [24] |
Moyers BA, Zhang JZ. Phylostratigraphic bias creates spurious patterns of genome evolution. Mol Biol Evol, 2015, 32(1): 258-267.
doi: 10.1093/molbev/msu286 pmid: 25312911 |
| [25] |
Moyers BA, Zhang JZ. Evaluating phylostratigraphic evidence for widespread de novo gene birth in genome evolution. Mol Biol Evol, 2016, 33(5): 1245-1256.
doi: 10.1093/molbev/msw008 pmid: 26758516 |
| [26] |
Moyers BA, Zhang JZ. Further simulations and analyses demonstrate open problems of phylostratigraphy. Genome Biol Evol, 2017, 9(6): 1519-1527.
doi: 10.1093/gbe/evx109 pmid: 28637261 |
| [27] |
Domazet-Lošo T, Carvunis AR, Albà MM, Šestak MS, Bakaric R, Neme R, Tautz D. No evidence for phylostratigraphic bias impacting inferences on patterns of gene emergence and evolution. Mol Biol Evol, 2017, 34(4): 843-856.
doi: 10.1093/molbev/msw284 pmid: 28087778 |
| [28] |
Knowles DG, McLysaght A. Recent de novo origin of human protein-coding genes. Genome Res, 2009, 19(10): 1752-1759.
doi: 10.1101/gr.095026.109 pmid: 19726446 |
| [29] | Xie C, Zhang YE, Chen JY, Liu CJ, Zhou WZ, Li Y, Zhang M, Zhang RL, Wei LP, Li CY. Hominoid-specific de novo protein-coding genes originating from long non-coding RNAs. PLoS Genet, 2012, 8(9): e1002942. |
| [30] | Murphy DN, McLysaght A. De novo origin of protein- coding genes in murine rodents. PLoS One, 2012, 7(11): e48650. |
| [31] |
Chen L, DeVries AL, Cheng CH. Evolution of antifreeze glycoprotein gene from a trypsinogen gene in Antarctic notothenioid fish. Proc Natl Acad Sci USA, 1997, 94(8): 3811-3816.
pmid: 9108060 |
| [32] | Cai J, Zhao RP, Jiang HF, Wang W. De novo origination of a new protein-coding gene in Saccharomyces cerevisiae. Genetics, 2008, 179(1): 487-496. |
| [33] |
Zhang L, Ren Y, Yang T, Li GW, Chen JH, Gschwend AR, Yu Y, Hou GX, Zi J, Zhou R, Wen B, Zhang JW, Chougule K, Wang MH, Copetti D, Peng ZY, Zhang CJ, Zhang Y, Ouyang YD, Wing RA, Liu SQ, Long MY. Rapid evolution of protein diversity by de novo origination in Oryza. Nat Ecol Evol, 2019, 3(4): 679-690.
doi: 10.1038/s41559-019-0822-5 pmid: 30858588 |
| [34] |
Kaessmann H. Origins, evolution, and phenotypic impact of new genes. Genome Res, 2010, 20(10): 1313-1326.
doi: 10.1101/gr.101386.109 pmid: 20651121 |
| [35] | Cui X, Lv Y, Chen ML, Nikoloski Z, Twell D, Zhang DB. Young genes out of the male: an insight from evolutionary age analysis of the pollen transcriptome. Mol Plant, 2015, 8(6): 935-945. |
| [36] | Witt E, Benjamin S, Svetec N, Zhao L. Testis single-cell RNA-seq reveals the dynamics of de novo gene transcription and germline mutational bias in Drosophila. eLife, 2019, 8: e47138. |
| [37] |
Levine MT, Jones CD, Kern AD, Lindfors HA, Begun DJ. Novel genes derived from noncoding DNA in Drosophila melanogaster are frequently X-linked and exhibit testis-biased expression. Proc Natl Acad Sci USA, 2006, 103(26): 9935-9939.
doi: 10.1073/pnas.0509809103 pmid: 16777968 |
| [38] | Begun DJ, Lindfors HA, Kern AD, Jones CD. Evidence for de novo evolution of testis-expressed genes in the Drosophila yakuba/Drosophila erecta clade. Genetics, 2007, 176(2): 1131-1137. |
| [39] |
Vibranovski MD, Zhang Y, Long MY. General gene movement off the X chromosome in the Drosophila genus. Genome Res, 2009, 19(5): 897-903.
doi: 10.1101/gr.088609.108 pmid: 19251740 |
| [40] |
Gubala AM, Schmitz JF, Kearns MJ, Vinh TT, Bornberg-Bauer E, Wolfner MF, Findlay GD. The goddard and saturn genes are essential for Drosophila male fertility and may have arisen de novo. Mol Biol Evol, 2017, 34(5): 1066-1082.
doi: 10.1093/molbev/msx057 pmid: 28104747 |
| [41] | Reinhardt JA, Wanjiru BM, Brant AT, Saelao P, Begun DJ, Jones CD. De novo ORFs in Drosophila are important to organismal fitness and evolved rapidly from previously non-coding sequences. PLoS Genet, 2013, 9(10): e1003860. |
| [42] |
Heinen TJAJ, Staubach F, Häming D, Tautz D. Emergence of a new gene from an intergenic region. Curr Biol, 2009, 19(18): 1527-1531.
doi: 10.1016/j.cub.2009.07.049 pmid: 19733073 |
| [43] | Xie C, Bekpen C, Künzel S, Keshavarz M, Krebs-Wheaton R, Skrabar N, Ullrich KK, Tautz D. A de novo evolved gene in the house mouse regulates female pregnancy cycles. eLife, 2019, 8: e44392. |
| [44] | Kleene KC. A possible meiotic function of the peculiar patterns of gene expression in mammalian spermatogenic cells. Mech Dev, 2001, 106(1-2): 3-23. |
| [45] |
Kleene KC. Sexual selection, genetic conflict, selfish genes, and the atypical patterns of gene expression in spermatogenic cells. Dev Biol, 2005, 277(1): 16-26.
doi: 10.1016/j.ydbio.2004.09.031 pmid: 15572136 |
| [46] | Green EW, Fedele G, Giorgini F, Kyriacou CP. A Drosophila RNAi collection is subject to dominant phenotypic effects. Nat Methods, 2014, 11(3): 222-223. |
| [47] | Kondo S, Vedanayagam J, Mohammed J, Eizadshenass S, Kan LJ, Pang N, Aradhya R, Siepel A, Steinhauer J, Lai EC. New genes often acquire male-specific functions but rarely become essential in Drosophila. Genes Dev, 2017, 31(18): 1841-1846. |
| [48] | Xia SQ, VanKuren NW, Chen CY, Zhang L, Kemkemer C, Shao Y, Jia HX, Lee U, Advani AS, Gschwend A, Vibranovski MD, Chen SD, Zhang YE, Long MY. Genomic analyses of new genes and their phenotypic effects reveal rapid evolution of essential functions in Drosophila development. PLoS Genet, 2021, 17(7): e1009654. |
| [49] |
Emerson JJ, Kaessmann H, Betrán E, Long MY. Extensive gene traffic on the mammalian X chromosome. Science, 2004, 303(5657): 537-540.
pmid: 14739461 |
| [50] |
Zhao Y, Hawes J, Popov KM, Jaskiewicz J, Shimomura Y, Crabb DW, Harris RA. Site-directed mutagenesis of phosphorylation sites of the branched chain alpha-ketoacid dehydrogenase complex. J Biol Chem, 1994, 269(28): 18583-18587.
pmid: 8034607 |
| [51] |
Wildman DE, Uddin M, Liu GZ, Grossman LI, Goodman M. Implications of natural selection in shaping 99.4% nonsynonymous DNA identity between humans and chimpanzees: enlarging genus Homo. Proc Natl Acad Sci USA, 2003, 100(12): 7181-7188.
doi: 10.1073/pnas.1232172100 pmid: 12766228 |
| [52] | An NA, Zhang J, Mo F, Luan XK, Tian L, Shen QS, Li XS, Li CQ, Zhou FQ, Zhang BY, Ji MJ, Qi JH, Zhou WZ, Ding WQ, Chen JY, Yu J, Zhang L, Shu SK, Hu BY, Li CY. De novo genes with an lncRNA origin encode unique human brain developmental functionality. Nat Ecol Evol, 2023, 7(2): 264-278. |
| [53] |
Rice WR, Chippindale AK. Sexual recombination and the power of natural selection. Science, 2001, 294(5542): 555-559.
pmid: 11641490 |
| [54] |
Boehm T, Hirano M, Holland SJ, Das S, Schorpp M, Cooper MD. Evolution of alternative adaptive immune systems in vertebrates. Annu Rev Immunol, 2018, 36: 19-42.
doi: 10.1146/annurev-immunol-042617-053028 pmid: 29144837 |
| [55] |
Böhm I, Schild H. Apoptosis: the complex scenario for a silent cell death. Mol Imaging Biol, 2003, 5(1): 2-14.
pmid: 14499155 |
| [56] | Janeway CA, Travers P, Walport M. Immunobiology:the Immune System in Health and Disease. 5th edition. New York: Garland Science, 2001. |
| [57] |
Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell, 2015, 161(5): 1187-1201.
doi: S0092-8674(15)00500-0 pmid: 26000487 |
| [58] | de Magãlhaes JP, Costa J. A database of vertebrate longevity records and their relation to other life-history traits. J Evol Biol, 2009, 22(8): 1770-1774. |
| [59] | Zhang L, Park JJ, Dong MB, Arsala D, Xia SQ, Chen JH, Sosa D, Atlas JE, Long MY, Chen SD. Human gene age dating reveals an early and rapid evolutionary construction of the adaptive immune system. Genome Biol Evol, 2023, 15(5): evad081. |
| [60] |
Cooper MD, Alder MN. The evolution of adaptive immune systems. Cell, 2006, 124(4): 815-822.
doi: 10.1016/j.cell.2006.02.001 pmid: 16497590 |
| [61] |
Smith JJ, Timoshevskaya N, Ye CX, Holt C, Keinath MC, Parker HJ, Cook ME, Hess JE, Narum SR, Lamanna F, Kaessmann H, Timoshevskiy VA, Waterbury CKM, Saraceno C, Wiedemann LM, Robb SMC, Baker C, Eichler EE, Hockman D, Sauka-Spengler T, Yandell M, Krumlauf R, Elgar G, Amemiya CT. The sea lamprey germline genome provides insights into programmed genome rearrangement and vertebrate evolution. Nat Genet, 2018, 50(2): 270-277.
doi: 10.1038/s41588-017-0036-1 pmid: 29358652 |
| [62] | Yu DQ, Ren YD, Uesaka M, Beavan AJS, Muffato M, Shen JY, Li YX, Sato I, Wan WT, Clark JW, Keating JN, Carlisle EM, Dearden RP, Giles S, Randle E, Sansom RS, Feuda R, Fleming JF, Sugahara F, Cummins C, Patricio M, Akanni W, D'Aniello S, Bertolucci C, Irie N, Alev C, Sheng GJ, de Mendoza A, Maeso I, Irimia M, Fromm B, Peterson KJ, Das S, Hirano M, Rast JP, Cooper MD, Paps J, Pisani D, Kuratani S, Martin FJ, Wang W, Donoghue PCJ, Zhang YE, Pascual-Anaya J. Hagfish genome elucidates vertebrate whole-genome duplication events and their evolutionary consequences. Nat Ecol Evol, 2024, 8(3): 519-535. |
| [1] | 陈晓晨,孙浩,米冬青,黄小琴,林克勤,易文,于亮,史磊,史荔,杨昭庆,褚嘉祐. 不同群体中ATXN2基因编码区CAG重复的变异研究[J]. 遗传, 2011, 33(4): 353-357. |
| [2] | 陈金宝,史磊,姚宇峰,史荔,于亮,林克勤,陶玉芬,易文,黄小琴,孙浩,褚嘉祐. 云南傣族、汉族HLA-G基因14 bp插入/缺失多态性研究[J]. 遗传, 2010, 32(6): 577-582. |
| [3] | 林栲,李海鹏. DNA水平上检测正选择方法的研究进展[J]. 遗传, 2009, 31(9): 896-902. |
| [4] | 顾明亮,汪业军,史磊,张永彪,褚嘉祐. 中国3个不同地域藏族群体线粒体ATP6、ATP8和Cyt b基因的比较: 探查自然选择在基因组的印记 [J]. 遗传, 2009, 31(2): 147-152. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||

www.chinagene.cn
备案号:京ICP备09063187号-4
总访问:,今日访问:,当前在线:

