遗传 ›› 2025, Vol. 47 ›› Issue (2): 183-199.doi: 10.16288/j.yczz.24-212
收稿日期:2024-07-18
修回日期:2024-09-02
出版日期:2025-02-20
发布日期:2024-10-11
通讯作者:
周琦,博士,教授,研究方向:性染色体演化、性别决定和性腺发育。E-mail: zhouqi1982@zju.edu.cn作者简介:刘靖,博士,研究方向:染色体演化。E-mail: liujing1993@zju.edu.cn
基金资助:Received:2024-07-18
Revised:2024-09-02
Published:2025-02-20
Online:2024-10-11
Supported by:摘要:
染色体作为位于细胞核中的基本遗传物质单位,在真核生物的演化历史中经历了广泛和复杂变化,很多变化的模式和机制在各种包括癌症在内的疾病中拥有共性。但很长时间内生物学家只能局限于基于荧光原位杂交技术等分辨率比较低的研究手段。当前日新月异的高通量测序技术正在快速革新人们对不同物种、同一物种不同个体甚至是单个个体不同细胞水平染色体变化的认识。本文聚焦于脊椎动物的染色体演化,概述了染色体重排在物种形成中的作用,驱动染色体重排的分子机制,祖先染色体到现今染色体的演变模式,以及性染色体作为研究染色体演化的一般范式意义,最后讨论了合成生物学为染色体演化研究领域带来的新机遇和挑战,以期为理解和研究脊椎动物的染色体演化提供新的见解和参考。
刘靖, 周琦. 脊椎动物染色体在序列和空间构象的演化[J]. 遗传, 2025, 47(2): 183-199.
Jing Liu, Qi Zhou. The evolution of sequences and spatial conformation in vertebrate chromosomes[J]. Hereditas(Beijing), 2025, 47(2): 183-199.
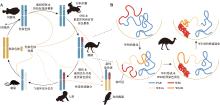
图4
脊椎动物的性染色体演化 A:性染色体转换(turn-over)(图根据参考文献[95]修改)。性染色体在演化过程中会经历逐步的退化阶段,分别由代表性的物种标示;性染色体转换可以避开“演化陷阱”,然而处于高度退化阶段的性染色体很难再发生转换,但可以通过染色体融合形成新的性染色体;B:鸸鹋W染色体在三维空间的演变模式(图根据参考文献[39]修改)。鸸鹋的性染色体大部分是同质性的,只有1/3左右的区域发生了退化,分为两个演化断层:S0和S1。Hi-C和基因组证据提示了鸸鹋W染色体的退化模式:WS0退化并高度异染色质化,空间上靠近核膜;WS1可能由一次倒位事件产生,并形成了WS1A和WS1B两部分,后者可能由于WS0异染色质的的邻近扩散效应使得WS1B相比WS1A发生了更快速地退化。"

| [1] |
O’Brien SJ, Nash WG. Genetic mapping in mammals: chromosome map of domestic cat. Science, 1982, 216(4543): 257-265.
pmid: 7063884 |
| [2] |
Sawyer JR, Hozier JC. High resolution of mouse chromosomes: banding conservation between man and mouse. Science, 1986, 232(4758): 1632-1635.
pmid: 3715469 |
| [3] |
Murphy WJ, Bourque G, Tesler G, Pevzner P, O’Brien SJ. Reconstructing the genomic architecture of mammalian ancestors using multispecies comparative maps. Hum Genomics, 2003, 1(1): 30-40.
doi: 10.1186/1479-7364-1-1-30 pmid: 15601531 |
| [4] |
Griffin DK, Robertson LBW, Tempest HG, Skinner BM. The evolution of the avian genome as revealed by comparative molecular cytogenetics. Cytogenet Genome Res, 2007, 117(1-4): 64-77.
doi: 10.1159/000103166 pmid: 17675846 |
| [5] |
Ruiz-Herrera A, Farré M, Robinson TJ. Molecular cytogenetic and genomic insights into chromosomal evolution. Heredity, 2012, 108(1): 28-36.
doi: 10.1038/hdy.2011.102 pmid: 22108627 |
| [6] |
Richard F, Lombard M, Dutrillaux B. Reconstruction of the ancestral karyotype of eutherian mammals. Chromosome Res, 2003, 11(6): 605-618.
doi: 10.1023/a:1024957002755 pmid: 14516069 |
| [7] |
Blanchette M, Green ED, Miller W, Haussler D. Reconstructing large regions of an ancestral mammalian genome in silico. Genome Res, 2004, 14(12): 2412-2423.
doi: 10.1101/gr.2800104 pmid: 15574820 |
| [8] |
Kohn M, Högel J, Vogel W, Minich P, Kehrer-Sawatzki H, Graves JAM, Hameister H. Reconstruction of a 450-My-old ancestral vertebrate protokaryotype. Trends Genet, 2006, 22(4): 203-210.
pmid: 16517001 |
| [9] |
Lin CC, Sasi R, Fan YS, Chen ZQ. New evidence for tandem chromosome fusions in the karyotypic evolution of Asian muntjacs. Chromosoma, 1991, 101(1): 19-24.
doi: 10.1007/BF00360682 pmid: 1769270 |
| [10] |
Gallardo MH, González CA, Cebrián I. Molecular cytogenetics and allotetraploidy in the red vizcacha rat, Tympanoctomys barrerae (Rodentia, Octodontidae). Genomics, 2006, 88(2): 214-221.
pmid: 16580173 |
| [11] |
Sturtevant AH. A case of rearrangement of genes in Drosophila. Proc Natl Acad Sci USA, 1921, 7(8): 235-237.
pmid: 16576597 |
| [12] | Dobzhansky T, Gould SJ. Genetics and the Origin of Species. Columbia University Press, 1982. |
| [13] |
Rieseberg LH. Chromosomal rearrangements and speciation. Trends Ecol Evol, 2001, 16(7): 351-358.
doi: 10.1016/s0169-5347(01)02187-5 pmid: 11403867 |
| [14] | Lowry DB, Willis JH. A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLoS Biol, 2010, 8(9): e1000500. |
| [15] | Fuller ZL, Koury SA, Phadnis N, Schaeffer SW. How chromosomal rearrangements shape adaptation and speciation: case studies in Drosophila pseudoobscura and its sibling species Drosophila persimilis. Mol Ecol, 2019, 28(6): 1283-1301. |
| [16] |
Kocher TD. Adaptive evolution and explosive speciation: the cichlid fish model. Nat Rev Genet, 2004, 5(4): 288-298.
doi: 10.1038/nrg1316 pmid: 15131652 |
| [17] | Jones FC, Grabherr MG, Chan YF, Russell P, Mauceli E, Johnson J, Swofford R, Pirun M, Zody MC, White S, Birney E, Searle S, Schmutz J, Grimwood J, Dickson MC, Myers RM, Miller CT, Summers BR, Knecht AK, Brady SD, Zhang H, Pollen AA, Howes T, Amemiya C, Broad Institute Genome Sequencing Platform & Whole Genome Assembly Team, Baldwin J, Bloom T, Jaffe DB, Nicol R, Wilkinson J, Lander ES, Di Palma F, Lindblad-Toh K, Kingsley DM. The genomic basis of adaptive evolution in threespine sticklebacks. Nature, 2012, 484(7392): 55-61. |
| [18] |
Potter S, Bragg JG, Blom MPK, Deakin JE, Kirkpatrick M, Eldridge MDB, Moritz C. Chromosomal speciation in the genomics era: disentangling phylogenetic evolution of rock-wallabies. Front Genet, 2017, 8: 10.
doi: 10.3389/fgene.2017.00010 pmid: 28265284 |
| [19] |
Farré M, Micheletti D, Ruiz-Herrera A. Recombination rates and genomic shuffling in human and chimpanzee--a new twist in the chromosomal speciation theory. Mol Biol Evol, 2013, 30(4): 853-864.
doi: 10.1093/molbev/mss272 pmid: 23204393 |
| [20] |
Tubbs A, Nussenzweig A. Endogenous dna damage as a source of genomic instability in cancer. Cell, 2017, 168(4): 644-656.
doi: S0092-8674(17)30005-3 pmid: 28187286 |
| [21] | Jasin M, Rothstein R. Repair of strand breaks by homologous recombination. Cold Spring Harb Perspect Biol, 2013, 5(11): a012740. |
| [22] |
Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem, 2010, 79: 181-211.
doi: 10.1146/annurev.biochem.052308.093131 pmid: 20192759 |
| [23] | Misteli T, Soutoglou E. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat Rev Mol Cell Biol, 2009, 10(4): 243-254. |
| [24] |
Jakob B, Splinter J, Durante M, Taucher-Scholz G. Live cell microscopy analysis of radiation-induced DNA double-strand break motion. Proc Natl Acad Sci USA, 2009, 106(9): 3172-3177.
doi: 10.1073/pnas.0810987106 pmid: 19221031 |
| [25] |
Lukásová E, Kozubek S, Kozubek M, Kjeronská J, Rýznar L, Horáková J, Horneck G. Localisation and distance between ABL and BCR genes in interphase nuclei of bone marrow cells of control donors and patients with chronic myeloid leukaemia. Hum Genet, 1997, 100(5-6): 525-535.
pmid: 9341866 |
| [26] |
Roix JJ, McQueen PG, Munson PJ, Parada LA, Misteli T. Spatial proximity of translocation-prone gene loci in human lymphomas. Nat Genet, 2003, 34(3): 287-291.
doi: 10.1038/ng1177 pmid: 12808455 |
| [27] |
Zhang Y, McCord RP, Ho YJ, Lajoie BR, Hildebrand DG, Simon AC, Becker MS, Alt FW, Dekker J. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell, 2012, 148(5): 908-921.
doi: 10.1016/j.cell.2012.02.002 pmid: 22341456 |
| [28] |
Nikiforova MN, Stringer JR, Blough R, Medvedovic M, Fagin JA, Nikiforov YE. Proximity of chromosomal loci that participate in radiation-induced rearrangements in human cells. Science, 2000, 290(5489): 138-141.
pmid: 11021799 |
| [29] | Cremer T, Cremer M. Chromosome territories. Cold Spring Harb Perspect Biol, 2010, 2(3): a003889. |
| [30] |
Misteli T. Beyond the sequence: cellular organization of genome function. Cell, 2007, 128(4): 787-800.
pmid: 17320514 |
| [31] |
Haber JE, Leung WY. Lack of chromosome territoriality in yeast: promiscuous rejoining of broken chromosome ends. Proc Natl Acad Sci USA, 1996, 93(24): 13949-13954.
pmid: 8943041 |
| [32] |
Hoencamp C, Dudchenko O, Elbatsh AMO, Brahmachari S, Raaijmakers JA, van Schaik T, Sedeño Cacciatore Á, Contessoto VG, van den Broek B, Mhaskar AN, Teunissen H, St Hilaire BG, Weisz D, Omer AD, Pham M, Colaric Z, Yang Z, Rao SSP, Mitra N, Lui C, Yao WJ, Khan R, Moroz LL, Kohn A, St Leger J, Mena A, Holcroft K, Gambetta MC, Lim F, Farley E, Stein N, Haddad A, Chauss D, Mutlu AS, Wang MC, Young ND, Hildebrandt E, Cheng HH, Knight CJ, Burnham TLU, Hovel KA, Beel AJ, Mattei PJ, Kornberg RD, Warren WC, Cary G, Gómez-Skarmeta JL, Hinman V, Lindblad-Toh K, Di Palma F, Maeshima K, Multani AS, Pathak S, Nel-Themaat L, Behringer RR, Kaur P, Medema RH, van Steensel B, de Wit E, Onuchic JN, Di Pierro M, Lieberman Aiden E, Rowland BD. 3D genomics across the tree of life reveals condensin II as a determinant of architecture type. Science, 2021, 372(6545): 984-989.
doi: 10.1126/science.abe2218 pmid: 34045355 |
| [33] | Rabl C. Über Zelltheilung. Morphologisches Jahrbuch, 1885, 10: 214-330. |
| [34] | Kitanishi Y, Sugishita H, Gotoh Y, Hirata Y. Changes of chromosomal architecture before establishment of chromosome territories revealed by recurrence plot reconstruction. bioRxiv, 2022, doi:10.1101/2021.05.20.444916. |
| [35] |
Bredeson JV, Mudd AB, Medina-Ruiz S, Mitros T, Smith OK, Miller KE, Lyons JB, Batra SS, Park J, Berkoff KC, Plott C, Grimwood J, Schmutz J, Aguirre-Figueroa G, Khokha MK, Lane M, Philipp I, Laslo M, Hanken J, Kerdivel G, Buisine N, Sachs LM, Buchholz DR, Kwon T, Smith-Parker H, Gridi-Papp M, Ryan MJ, Denton RD, Malone JH, Wallingford JB, Straight AF, Heald R, Hockemeyer D, Harland RM. Conserved chromatin and repetitive patterns reveal slow genome evolution in frogs. Nat Commun, 2024, 15(1): 579.
doi: 10.1038/s41467-023-43012-9 pmid: 38233380 |
| [36] |
Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA. Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol, 1999, 145(6): 1119-1131.
pmid: 10366586 |
| [37] |
Federico C, Scavo C, Cantarella CD, Motta S, Saccone S, Bernardi G. Gene-rich and gene-poor chromosomal regions have different locations in the interphase nuclei of cold-blooded vertebrates. Chromosoma, 2006, 115(2): 123-128.
pmid: 16404627 |
| [38] | Waters PD, Patel HR, Ruiz-Herrera A, Álvarez-González L, Lister NC, Simakov O, Ezaz T, Kaur P, Frere C, Grützner F, Georges A, Marshall Graves JA. Microchromosomes are building blocks of bird, reptile, and mammal chromosomes. Proc Natl Acad Sci USA, 2021, 118(45): e2112494118. |
| [39] |
Liu J, Wang ZJ, Li J, Xu LH, Liu JQ, Feng SH, Guo CX, Chen SC, Ren ZJ, Rao JP, Wei K, Chen YZ, Jarvis ED, Zhang GJ, Zhou Q. A new emu genome illuminates the evolution of genome configuration and nuclear architecture of avian chromosomes. Genome Res, 2021, 31(3): 497-511.
doi: 10.1101/gr.271569.120 pmid: 33408157 |
| [40] |
Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet, 2001, 2(4): 292-301.
doi: 10.1038/35066075 pmid: 11283701 |
| [41] |
Price BD, D’Andrea AD. Chromatin remodeling at DNA double-strand breaks. Cell, 2013, 152(6): 1344-1354.
doi: 10.1016/j.cell.2013.02.011 pmid: 23498941 |
| [42] | Aymard F, Legube G. A TAD closer to ATM. Mol Cell Oncol, 2016, 3(3): e1134411. |
| [43] | Habermann FA, Lacerda O, Sbalqueiro IJ, Wienberg J, Müller S. Chromosome reshuffling in birds of prey: the karyotype of the world’s largest eagle (Harpy eagle, Harpia harpyja) compared to that of the chicken (Gallus gallus). Chromosoma, 2005, 114(5): 338-343. |
| [44] |
Cournac A, Koszul R, Mozziconacci J. The 3D folding of metazoan genomes correlates with the association of similar repetitive elements. Nucleic Acids Res, 2016, 44(1): 245-255.
doi: 10.1093/nar/gkv1292 pmid: 26609133 |
| [45] |
Lu JY, Chang L, Li T, Wang T, Yin YF, Zhan G, Han X, Zhang K, Tao YB, Percharde M, Wang L, Peng Q, Yan PX, Zhang H, Bi XJ, Shao W, Hong YT, Wu ZY, Ma RZ, Wang PZ, Li WZ, Zhang J, Chang Z, Hou YP, Zhu B, Ramalho-Santos M, Li PL, Xie W, Na J, Sun YJ, Shen XH. Homotypic clustering of L1 and B1/Alu repeats compartmentalizes the 3D genome. Cell Res, 2021, 31(6): 613-630.
doi: 10.1038/s41422-020-00466-6 pmid: 33514913 |
| [46] |
Jarmuz-Szymczak M, Janiszewska J, Szyfter K, Shaffer LG. Narrowing the localization of the region breakpoint in most frequent Robertsonian translocations. Chromosome Res, 2014, 22(4): 517-532.
doi: 10.1007/s10577-014-9439-3 pmid: 25179263 |
| [47] | Guarracino A, Buonaiuto S, de Lima LG, Potapova T, Rhie A, Koren S, Rubinstein B, Fischer C, Human Pangenome Reference Consortium, Gerton JL, Phillippy AM, Colonna V, Garrison E. Recombination between heterologous human acrocentric chromosomes. Nature, 2023, 617(7960): 335-343. |
| [48] | Knutsen T, Padilla-Nash HM, Wangsa D, Barenboim-Stapleton L, Camps J, McNeil N, Difilippantonio MJ, Ried T. Definitive molecular cytogenetic characterization of 15 colorectal cancer cell lines. Genes Chromosomes Cancer, 2010, 49(3): 204-223. |
| [49] |
Yadav V, Sun S, Coelho MA, Heitman J. Centromere scission drives chromosome shuffling and reproductive isolation. Proc Natl Acad Sci USA, 2020, 117(14): 7917-7928.
doi: 10.1073/pnas.1918659117 pmid: 32193338 |
| [50] |
Yin Y, Fan HZ, Zhou BT, Hu YB, Fan GY, Wang JH, Zhou F, Nie WH, Zhang CZ, Liu L, Zhong ZY, Zhu WB, Liu GC, Lin ZS, Liu C, Zhou J, Huang GP, Li ZH, Yu JP, Zhang YL, Yang Y, Zhuo BZ, Zhang BW, Chang J, Qian HY, Peng YM, Chen XQ, Chen L, Li ZP, Zhou Q, Wang W, Wei FW. Molecular mechanisms and topological consequences of drastic chromosomal rearrangements of muntjac deer. Nat Commun, 2021, 12(1): 6858.
doi: 10.1038/s41467-021-27091-0 pmid: 34824214 |
| [51] | Gozashti L, Harringmeyer OS, Hoekstra HE. How repeats rearrange chromosomes in deer mice. bioRxiv, 2024, doi:10.1101/2024.05.29.596518. |
| [52] | Baalsrud HT, Garmann-Aarhus B, Enevoldsen ELG, Krabberød AK, Fischer D, Tooming-Klunderud A, Skage M, Árnyasi M, Sandve SR, Jakobsen K, Nielsen R, Boessenkool S, Tørresen OK. Evolutionary new centromeres in the snowy owl genome putatively seeded from a transposable element. bioRxiv, 2024, doi:10.1101/2024.07.05.602039. |
| [53] | Morris SC, Caron J-B. A primitive fish from the Cambrian of North America. Nature, 2014, 512(7515): 419-422. |
| [54] |
Miyashita T, Coates MI, Farrar R, Larson P, Manning PL, Wogelius RA, Edwards NP, Anné J, Bergmann U, Palmer AR, Currie PJ. Hagfish from the Cretaceous Tethys Sea and a reconciliation of the morphological-molecular conflict in early vertebrate phylogeny. Proc Natl Acad Sci USA, 2019, 116(6): 2146-2151.
doi: 10.1073/pnas.1814794116 pmid: 30670644 |
| [55] | Yu DQ, Ren YD, Uesaka M, Beavan AJS, Muffato M, Shen JY, Li YX, Sato I, Wan WT, Clark JW, Keating JN, Carlisle EM, Dearden RP, Giles S, Randle E, Sansom RS, Feuda R, Fleming JF, Sugahara F, Cummins C, Patricio M, Akanni W, D'Aniello S, Bertolucci C, Irie N, Alev C, Sheng GJ, de Mendoza A, Maeso I, Irimia M, Fromm B, Peterson KJ, Das S, Hirano M, Rast JP, Cooper MD, Paps J, Pisani D, Kuratani S, Martin FJ, Wang W, Donoghue PCJ, Zhang YE, Pascual-Anaya J. Hagfish genome elucidates vertebrate whole-genome duplication events and their evolutionary consequences. Nat Ecol Evol, 2024, 8(3): 519-535. |
| [56] | Marlétaz F, Timoshevskaya N, Timoshevskiy VA, Parey E, Simakov O, Gavriouchkina D, Suzuki M, Kubokawa K, Brenner S, Smith JJ, Rokhsar DS. The hagfish genome and the evolution of vertebrates. Nature, 2024, 627(8005): 811-820. |
| [57] |
Smith JJ, Antonacci F, Eichler EE, Amemiya CT. Programmed loss of millions of base pairs from a vertebrate genome. Proc Natl Acad Sci USA, 2009, 106(27): 11212-11217.
doi: 10.1073/pnas.0902358106 pmid: 19561299 |
| [58] | Ohno S. Evolution by Gene Duplication. Springer Berlin Heidelberg, 1970. |
| [59] | Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol, 2005, 3(10): e314. |
| [60] |
Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV, Jurka J, Genikhovich G, Grigoriev IV, Lucas SM, Steele RE, Finnerty JR, Technau U, Martindale MQ, Rokhsar DS. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science, 2007, 317(5834): 86-94.
doi: 10.1126/science.1139158 pmid: 17615350 |
| [61] | Huang Z, Xu LH, Cai C, Zhou YT, Liu J, Xu ZX, Zhu ZX, Kang W, Cen W, Pei SR, Chen D, Shi CG, Wu XT, Huang YJ, Xu CH, Yan YN, Yang Y, Xue T, He WJ, Hu XF, Zhang YD, Chen YQ, Bi CW, He CP, Xue LZ, Xiao SJ, Yue ZC, Jiang Y, Yu JK, Jarvis ED, Li G, Lin G, Zhang QJ, Zhou Q. Three amphioxus reference genomes reveal gene and chromosome evolution of chordates. Proc Natl Acad Sci USA, 2023, 120(10): e2201504120. |
| [62] |
Nakatani Y, Takeda H, Kohara Y, Morishita S. Reconstruction of the vertebrate ancestral genome reveals dynamic genome reorganization in early vertebrates. Genome Res, 2007, 17(9): 1254-1265.
doi: 10.1101/gr.6316407 pmid: 17652425 |
| [63] |
Sacerdot C, Louis A, Bon C, Berthelot C. Roest Crollius H. Chromosome evolution at the origin of the ancestral vertebrate genome. Genome Biol, 2018, 19(1): 166.
doi: 10.1186/s13059-018-1559-1 pmid: 30333059 |
| [64] | Putnam NH, Butts T, Ferrier DEK, Furlong RF, Hellsten U, Kawashima T, Robinson-Rechavi M, Shoguchi E, Terry A, Yu JK, Benito-Gutiérrez EL, Dubchak I, Garcia-Fernàndez J, Gibson-Brown JJ, Grigoriev IV, Horton AC, de Jong PJ, Jurka J, Kapitonov VV, Kohara Y, Kuroki Y, Lindquist E, Lucas S, Osoegawa K, Pennacchio LA, Salamov AA, Satou Y, Sauka-Spengler T, Schmutz J, Shin-I T, Toyoda A, Bronner-Fraser M, Fujiyama A, Holland LZ, Holland PW, Satoh N, Rokhsar DS. The amphioxus genome and the evolution of the chordate karyotype. Nature, 2008, 453(7198): 1064-1071. |
| [65] |
Simakov O, Marlétaz F, Yue J-X, O’Connell B, Jenkins J, Brandt A, Calef R, Tung CH, Huang TK, Schmutz J, Satoh N, Yu JK, Putnam NH, Green RE, Rokhsar DS. Deeply conserved synteny resolves early events in vertebrate evolution. Nat Ecol Evol, 2020, 4(6): 820-830.
doi: 10.1038/s41559-020-1156-z pmid: 32313176 |
| [66] | Nakatani Y, Shingate P, Ravi V, Pillai NE, Prasad A, McLysaght A, Venkatesh B. Reconstruction of proto- vertebrate, proto-cyclostome and proto-gnathostome genomes provides new insights into early vertebrate evolution. Nat Commun, 2021, 12(1): 4489. |
| [67] | Huang Z, Xu ZX, Bai H, Huang YJ, Kang N, Ding XT, Liu J, Luo HR, Yang CT, Chen WJ, Guo QX, Xue LZ, Zhang XP, Xu L, Chen ML, Fu HG, Chen YL, Yue ZC, Fukagawa T, Liu SL, Chang GB, Xu LH. Evolutionary analysis of a complete chicken genome. Proc Natl Acad Sci USA, 2023, 120(8): e2216641120. |
| [68] |
Wrigley JM, Graves JA. Karyotypic conservation in the mammalian order monotremata (subclass Prototheria). Chromosoma, 1988, 96(3): 231-247.
doi: 10.1007/BF00302363 pmid: 3359880 |
| [69] |
Braasch I, Gehrke AR, Smith JJ, Kawasaki K, Manousaki T, Pasquier J, Amores A, Desvignes T, Batzel P, Catchen J, Berlin AM, Campbell MS, Barrell D, Martin KJ, Mulley JF, Ravi V, Lee AP, Nakamura T, Chalopin D, Fan SH, Wcisel D, Cañestro C, Sydes J, Beaudry FE, Sun Y, Hertel J, Beam MJ, Fasold M, Ishiyama M, Johnson J, Kehr S, Lara M, Letaw JH, Litman GW, Litman RT, Mikami M, Ota T, Saha NR, Williams L, Stadler PF, Wang H, Taylor JS, Fontenot Q, Ferrara A, Searle SMJ, Aken B, Yandell M, Schneider I, Yoder JA, Volff JN, Meyer A, Amemiya CT, Venkatesh B, Holland PW, Guiguen Y, Bobe J, Shubin NH, Di Palma F, Alföldi J, Lindblad-Toh K, Postlethwait JH. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat Genet, 2016, 48(4): 427-437.
doi: 10.1038/ng.3526 pmid: 26950095 |
| [70] |
Feldman M, Levy AA, Fahima T, Korol A. Genomic asymmetry in allopolyploid plants: wheat as a model. J Exp Bot, 2012, 63(14): 5045-5059.
doi: 10.1093/jxb/ers192 pmid: 22859676 |
| [71] |
Fang L, Guan XY, Zhang TZ. Asymmetric evolution and domestication in allotetraploid cotton (Gossypium hirsutum L.). Crop J, 2017, 5(2): 159-165.
doi: 10.1016/j.cj.2016.07.001 |
| [72] | Zhang GJ, Li C, Li QY, Li B, Larkin DM, Lee C, Storz JF, Antunes A, Greenwold MJ, Meredith RW, Ödeen A, Cui J, Zhou Q, Xu LH, Pan HL, Wang ZJ, Jin LJ, Zhang P, Hu HF, Yang W, Hu J, Xiao J, Yang ZK, Liu Y, Xie QL, Yu H, Lian JM, Wen P, Zhang F, Li H, Zeng YL, Xiong ZJ, Liu SP, Zhou L, Huang ZY, An N, Wang J, Zheng QM, Xiong YQ, Wang GB, Wang B, Wang JJ, Fan Y, da Fonseca RR, Alfaro-Núñez A, Schubert M, Orlando L, Mourier T, Howard JT, Ganapathy G, Pfenning A, Whitney O, Rivas MV, Hara E, Smith J, Farré M, Narayan J, Slavov G, Romanov MN, Borges R, Borges R, Machado JP, Khan I, Springer MS, Gatesy J, Hoffmann FG, Opazo JC, Håstad O, Sawyer RH, Kim H, Kim KW, Kim HJ, Cho S, Li N, Huang YH, Bruford MW, Zhan XJ, Dixon A, Bertelsen MF, Derryberry E, Warren W, Wilson RK, Li SB, Ray DA, Green RE, O'Brien SJ, Griffin D, Johnson WE, Haussler D, Ryder OA, Willerslev E, Graves GR, Alström P, Fjeldså J, Mindell DP, Edwards SV, Braun EL, Rahbek C, Burt DW, Houde P, Zhang Y, Yang HM, Wang J, Avian Genome Consortium, Jarvis ED, Gilbert MT, Wang J. Comparative genomics reveals insights into avian genome evolution and adaptation. Science, 2014, 346(6215): 1311-1320. |
| [73] |
Schmidbaur H, Kawaguchi A, Clarence T, Fu X, Hoang OP, Zimmermann B, Ritschard EA, Weissenbacher A, Foster JS, Nyholm SV, Bates PA, Albertin CB, Tanaka E, Simakov O. Emergence of novel cephalopod gene regulation and expression through large-scale genome reorganization. Nat Commun, 2022, 13(1): 2172.
doi: 10.1038/s41467-022-29694-7 pmid: 35449136 |
| [74] |
Berthelot C, Muffato M, Abecassis J, Roest Crollius H. The 3D organization of chromatin explains evolutionary fragile genomic regions. Cell Rep, 2015, 10(11): 1913-1924.
pmid: 25801028 |
| [75] | Damas J, Kim J, Farré M, Griffin DK, Larkin DM. Reconstruction of avian ancestral karyotypes reveals differences in the evolutionary history of macro- and microchromosomes. Genome Biol, 2018, 19(1): 155. |
| [76] | Koepfli K P, Paten B, Genome 10K Community of Scientists, O’Brien SJ. The genome 10k project: a way forward. Annu Rev Anim Biosci, 2015, 3: 57-111. |
| [77] | Kim J, Farré M, Auvil L, Capitanu B, Larkin DM, Ma J, Lewin HA. Reconstruction and evolutionary history of eutherian chromosomes. Proc Natl Acad Sci USA, 2017, 114(27): E5379-E5388. |
| [78] | Damas J, Corbo M, Kim J, Turner-Maier J, Farré M, Larkin DM, Ryder OA, Steiner C, Houck ML, Hall S, Shiue L, Thomas S, Swale T, Daly M, Korlach J, Uliano-Silva M, Mazzoni CJ, Birren BW, Genereux DP, Johnson J, Lindblad-Toh K, Karlsson EK, Nweeia MT, Johnson RN, Zoonomia Consortium, Lewin HA. Evolution of the ancestral mammalian karyotype and syntenic regions. Proc Natl Acad Sci USA, 2022, 119(40): e2209139119. |
| [79] |
Feuk L. Inversion variants in the human genome: role in disease and genome architecture. Genome Med, 2010, 2(2): 11.
doi: 10.1186/gm132 pmid: 20156332 |
| [80] | Nomura T, Suzuki S, Miyauchi T, Takeda M, Shinkuma S, Fujita Y, Nishie W, Akiyama M, Shimizu H. Chromosomal inversions as a hidden disease-modifying factor for somatic recombination phenotypes. JCI Insight, 2018, 3(6): e97595. |
| [81] | O’Connor RE, Romanov MN, Kiazim LG, Barrett PM, Farré M, Damas J, Ferguson-Smith M, Valenzuela N, Larkin DM, Griffin DK. Reconstruction of the diapsid ancestral genome permits chromosome evolution tracing in avian and non-avian dinosaurs. Nat Commun, 2018, 9(1): 1883. |
| [82] | Zhou Y, Shearwin-Whyatt L, Li J, Song ZZ, Hayakawa T, Stevens D, Fenelon JC, Peel E, Cheng YY, Pajpach F, Bradley N, Suzuki H, Nikaido M, Damas J, Daish T, Perry T, Zhu ZX, Geng YC, Rhie A, Sims Y, Wood J, Haase B, Mountcastle J, Fedrigo O, Li QY, Yang HM, Wang J, Johnston SD, Phillippy AM, Howe K, Jarvis ED, Ryder OA, Kaessmann H, Donnelly P, Korlach J, Lewin HA, Graves J, Belov K, Renfree MB, Grutzner F, Zhou Q, Zhang GJ. Platypus and echidna genomes reveal mammalian biology and evolution. Nature, 2021, 592(7856): 756-762. |
| [83] | Nakatani Y, McLysaght A. Genomes as documents of evolutionary history: a probabilistic macrosynteny model for the reconstruction of ancestral genomes. Bioinformatics, 2017, 33(14): i369-i378. |
| [84] |
Parey E, Louis A, Montfort J, Guiguen Y, Roest Crollius H, Berthelot C. An atlas of fish genome evolution reveals delayed rediploidization following the teleost whole-genome duplication. Genome Res, 2022, 32(9): 1685-1697.
doi: 10.1101/gr.276953.122 pmid: 35961774 |
| [85] |
Garsmeur O, Schnable JC, Almeida A, Jourda C, D’Hont A, Freeling M. Two evolutionarily distinct classes of paleopolyploidy. Mol Biol Evol, 2014, 31(2): 448-454.
doi: 10.1093/molbev/mst230 pmid: 24296661 |
| [86] | Pyron RA, Pirro S, Hains T, Colston TJ, Myers EA, O’Connell KA, Beamer DA. The draft genome sequences of 50 salamander species (Caudata, Amphibia). Biodivers Genomes, 2024. doi:10.56179/001c.116891. |
| [87] | Ohno S. Sex Chromosomes and Sex-Linked Genes. Springer Berlin Heidelberg, 1966. |
| [88] | Stevens NM. Studies in Spermatogenesis: With Especial Reference to the Accessory Chromosome. Carnegie Institution of Washington, LLC, 1905. |
| [89] |
Meisel RP, Connallon T. The faster-X effect: integrating theory and data. Trends Genet, 2013, 29(9): 537-544.
doi: 10.1016/j.tig.2013.05.009 pmid: 23790324 |
| [90] | Kayserili MA, Gerrard DT, Tomancak P, Kalinka AT. An excess of gene expression divergence on the X chromosome in Drosophila embryos: implications for the faster-X hypothesis. PLoS Genet, 2012, 8(12): e1003200. |
| [91] | Muller HJ. The relation of recombination to mutational advance. Mutat Res, 1964, 106: 2-9. |
| [92] | Rice WR. Evolution of the Y sex chromosome in animals: Y chromosomes evolve through the degeneration of autosomes. Bioscience, 1996, 46(5): 331-343. |
| [93] |
Graves JAM. Sex chromosome specialization and degeneration in mammals. Cell, 2006, 124(5): 901-914.
pmid: 16530039 |
| [94] | Terao M, Ogawa Y, Takada S, Kajitani R, Okuno M, Mochimaru Y, Matsuoka K, Itoh T, Toyoda A, Kono T, Jogahara T, Mizushima S, Kuroiwa A. Turnover of mammal sex chromosomes in the Sry-deficient Amami spiny rat is due to male-specific upregulation of Sox9. Proc Natl Acad Sci USA, 2022, 119(49): e2211574119. |
| [95] |
Furman BLS, Metzger DCH, Darolti I, Wright AE, Sandkam BA, Almeida P, Shu JJ, Mank JE. Sex chromosome evolution: so many exceptions to the rules. Genome Biol Evol, 2020, 12(6): 750-763.
doi: 10.1093/gbe/evaa081 pmid: 32315410 |
| [96] | Kratochvíl L, Gamble T, Rovatsos M. Sex chromosome evolution among amniotes: is the origin of sex chromosomes non-random? Philos Trans R Soc Lond B Biol Sci, 2021, 376(1833): 20200108. |
| [97] | Myosho T, Takehana Y, Hamaguchi S, Sakaizumi M. Turnover of sex chromosomes in celebensis group medaka fishes. G3 (Bethesda), 2015, 5(12): 2685-2691. |
| [98] |
Jeffries DL, Lavanchy G, Sermier R, Sredl MJ, Miura I, Borzée A, Barrow LN, Canestrelli D, Crochet PA, Dufresnes C, Fu JZ, Ma WJ, Garcia CM, Ghali K, Nicieza AG, O'Donnell RP, Rodrigues N, Romano A, Martínez-Solano Í, Stepanyan I, Zumbach S, Brelsford A, Perrin N. A rapid rate of sex-chromosome turnover and non-random transitions in true frogs. Nat Commun, 2018, 9(1): 4088.
doi: 10.1038/s41467-018-06517-2 pmid: 30291233 |
| [99] |
Gamble T, Coryell J, Ezaz T, Lynch J, Scantlebury DP, Zarkower D. Restriction site-associated DNA sequencing (RAD-seq) reveals an extraordinary number of transitions among gecko sex-determining systems. Mol Biol Evol, 2015, 32(5): 1296-1309.
doi: 10.1093/molbev/msv023 pmid: 25657328 |
| [100] | Pokorná M, Kratochvíl L. Phylogeny of sex-determining mechanisms in squamate reptiles: are sex chromosomes an evolutionary trap? Zool J Linn Soc, 2009, 156(1): 168-183. |
| [101] | McKee BD, Yan RH, Tsai JH. Meiosis in male Drosophila. Spermatogenesis, 2012, 2(3): 167-184. |
| [102] | Berset-Brändli L, Jaquiéry J, Broquet T, Ulrich Y, Perrin N. Extreme heterochiasmy and nascent sex chromosomes in European tree frogs. Proc Biol Sci, 2008, 275(1642): 1577-1585. |
| [103] | Sardell JM, Cheng CD, Dagilis AJ, Ishikawa A, Kitano J, Peichel CL, Kirkpatrick M. Sex differences in recombination in sticklebacks. G3 (Bethesda), 2018, 8(6): 1971-1983. |
| [104] |
Bergero R, Gardner J, Bader B, Yong L, Charlesworth D. Exaggerated heterochiasmy in a fish with sex-linked male coloration polymorphisms. Proc Natl Acad Sci USA, 2019, 116(14): 6924-6931.
doi: 10.1073/pnas.1818486116 pmid: 30894479 |
| [105] |
Steinemann M, Steinemann S. Enigma of Y chromosome degeneration: neo-Y and neo-X chromosomes of Drosophila miranda a model for sex chromosome evolution. Genetica, 1998, 102-103(1-6): 409-420.
pmid: 9720292 |
| [106] | Bachtrog D, Hom E, Wong KM, Maside X, de Jong P.. Genomic degradation of a young Y chromosome in Drosophila miranda. Genome Biol, 2008, 9(2): R30. |
| [107] | Mongue AJ, Nguyen P, Voleníková A, Walters JR. Neo-sex chromosomes in the monarch butterfly, Danaus plexippus. G3 (Bethesda), 2017, 7(10): 3281-3294. |
| [108] | Huang Z, De O Furo I, Liu J, Peona V, Gomes AJB, Cen W, Huang H, Zhang YD, Chen D, Xue T, Zhang QJ, Yue ZC, Wang QX, Yu LY, Chen YL, Suh A, Xu LH. Recurrent chromosome reshuffling and the evolution of neo-sex chromosomes in parrots. Nat Commun, 2022, 13(1): 944. |
| [109] | Zhou Q, Wang J, Huang L, Nie WH, Wang JH, Liu Y, Zhao XY, Yang FT, Wang W. Neo-sex chromosomes in the black muntjac recapitulate incipient evolution of mammalian sex chromosomes. Genome Biol, 2008, 9: R98. |
| [110] |
Xu LL, Ren YD, Wu JH, Cui TT, Dong R, Huang C, Feng Z, Zhang TM, Yang P, Yuan JQ, Xu X, Liu J, Wang JH, Chen W, Mi D, Irwin DM, Yan YP, Xu LH, Yu XP, Li G. Evolution and expression patterns of the neo-sex chromosomes of the crested ibis. Nat Commun, 2024, 15(1): 1670.
doi: 10.1038/s41467-024-46052-x pmid: 38395916 |
| [111] |
Soh YQS, Alföldi J, Pyntikova T, Brown LG, Graves T, Minx PJ, Fulton RS, Kremitzki C, Koutseva N, Mueller JL, Rozen S, Hughes JF, Owens E, Womack JE, Murphy WJ, Cao Q, de Jong P, Warren WC, Wilson RK, Skaletsky H, Page DC. Sequencing the mouse Y chromosome reveals convergent gene acquisition and amplification on both sex chromosomes. Cell, 2014, 159(4): 800-813.
doi: 10.1016/j.cell.2014.09.052 pmid: 25417157 |
| [112] | Rhie A, Nurk S, Cechova M, Hoyt SJ, Taylor DJ, Altemose N, Hook PW, Koren S, Rautiainen M, Alexandrov IA, Allen J, Asri M, Bzikadze AV, Chen NC, Chin CS, Diekhans M, Flicek P, Formenti G, Fungtammasan A, Garcia Giron C, Garrison E, Gershman A, Gerton JL, Grady PGS, Guarracino A, Haggerty L, Halabian R, Hansen NF, Harris R, Hartley GA, Harvey WT, Haukness M, Heinz J, Hourlier T, Hubley RM, Hunt SE, Hwang S, Jain M, Kesharwani RK, Lewis AP, Li H, Logsdon GA, Lucas JK, Makalowski W, Markovic C, Martin FJ, Mc Cartney AM, McCoy RC, McDaniel J, McNulty BM, Medvedev P, Mikheenko A, Munson KM, Murphy TD, Olsen HE, Olson ND, Paulin LF, Porubsky D, Potapova T, Ryabov F, Salzberg SL, Sauria MEG, Sedlazeck FJ, Shafin K, Shepelev VA, Shumate A, Storer JM, Surapaneni L, Taravella Oill AM, Thibaud-Nissen F, Timp W, Tomaszkiewicz M, Vollger MR, Walenz BP, Watwood AC, Weissensteiner MH, Wenger AM, Wilson MA, Zarate S, Zhu YM, Zook JM, Eichler EE, O'Neill RJ, Schatz MC, Miga KH, Makova KD, Phillippy AM. The complete sequence of a human Y chromosome. Nature, 2023, 621(7978): 344-354. |
| [113] |
Malik P, Korfali N, Srsen V, Lazou V, Batrakou DG, Zuleger N, Kavanagh DM, Wilkie GS, Goldberg MW, Schirmer EC. Cell-specific and lamin-dependent targeting of novel transmembrane proteins in the nuclear envelope. Cell Mol Life Sci, 2010, 67(8): 1353-1369.
doi: 10.1007/s00018-010-0257-2 pmid: 20091084 |
| [114] |
Wang ZJ, Zhang JL, Xu XM, Witt C, Deng Y, Chen GJ, Meng GL, Feng SH, Xu LH, Szekely T, Zhang GJ, Zhou Q. Phylogeny and sex chromosome evolution of palaeognathae. J Genet Genomics, 2021, 49(2): 109-119.
doi: 10.1016/j.jgg.2021.06.013 pmid: 34872841 |
| [115] |
Shah PP, Keough KC, Gjoni K, Santini GT, Abdill RJ, Wickramasinghe NM, Dundes CE, Karnay A, Chen A, Salomon REA, Walsh PJ, Nguyen SC, Whalen S, Joyce EF, Loh KM, Dubois N, Pollard KS, Jain R. An atlas of lamina-associated chromatin across twelve human cell types reveals an intermediate chromatin subtype. Genome Biol, 2023, 24(1): 16.
doi: 10.1186/s13059-023-02849-5 pmid: 36691074 |
| [116] |
Peng T, Hou YP, Meng HW, Cao Y, Wang XT, Jia LM, Chen Q, Zheng Y, Sun YJ, Chen HB, Li TT, Li C. Mapping nucleolus-associated chromatin interactions using nucleolus Hi-C reveals pattern of heterochromatin interactions. Nat Commun, 2023, 14(1): 350.
doi: 10.1038/s41467-023-36021-1 pmid: 36681699 |
| [117] |
Hoffmann AA, Rieseberg LH. Revisiting the impact of inversions in evolution: from population genetic markers to drivers of adaptive shifts and speciation? Annu Rev Ecol Evol Syst, 2008, 39: 21-42.
pmid: 20419035 |
| [118] |
Nishikawa H, Iijima T, Kajitani R, Yamaguchi J, Ando T, Suzuki Y, Sugano S, Fujiyama A, Kosugi S, Hirakawa H, Tabata S, Ozaki K, Morimoto H, Ihara K, Obara M, Hori H, Itoh T, Fujiwara H. A genetic mechanism for female-limited Batesian mimicry in Papilio butterfly. Nat Genet, 2015, 47(4): 405-409.
doi: 10.1038/ng.3241 pmid: 25751626 |
| [119] | Hoff SNK, Maurstad M, Tørresen OK, Berg PR, Præbel K, Jakobsen KS, Jentoft S. Chromosomal fusions and large-scale inversions are key features for adaptation in Arctic codfish species. bioRxiv, 2024. doi:10.1101/2024.06.28.599280. |
| [120] | Kang ZJ, Liu YF, Xu LZ, Long ZJ, Huang D, Yang Y, Liu B, Feng JX, Pan YJ, Yan JS, Liu QT. The Philadelphia chromosome in leukemogenesis. Chin J Cancer, 2016, 35: 48. |
| [121] |
Zhang XM, Yan M, Yang ZH, Xiang H, Tang W, Cai XD, Wu QG, Liu X, Pei G, Li JS. Creation of artificial karyotypes in mice reveals robustness of genome organization. Cell Res, 2022, 32(11): 1026-1029.
doi: 10.1038/s41422-022-00722-x pmid: 36127403 |
| [122] |
Wang LB, Li ZK, Wang LY, Xu K, Ji TT, Mao YH, Ma SN, Liu T, Tu CF, Zhao Q, Fan XN, Liu C, Wang LY, Shu YJ, Yang N, Zhou Q, Li W. A sustainable mouse karyotype created by programmed chromosome fusion. Science, 2022, 377(6609): 967-975.
doi: 10.1126/science.abm1964 pmid: 36007034 |
| [123] | Luo JC, Sun XJ, Cormack BP, Boeke JD. Karyotype engineering by chromosome fusion leads to reproductive isolation in yeast. Nature, 2018, 560(7718): 392-396. |
| [124] |
Nurk S, Koren S, Rhie A, Rautiainen M, Bzikadze AV, Mikheenko A, Vollger MR, Altemose N, Uralsky L, Gershman A, Aganezov S, Hoyt SJ, Diekhans M, Logsdon GA, Alonge M, Antonarakis SE, Borchers M, Bouffard GG, Brooks SY, Caldas GV, Chen NC, Cheng HY, Chin CS, Chow W, de Lima LG, Dishuck PC, Durbin R, Dvorkina T, Fiddes IT, Formenti G, Fulton RS, Fungtammasan A, Garrison E, Grady PGS, Graves- Lindsay TA, Hall IM, Hansen NF, Hartley GA, Haukness M, Howe K, Hunkapiller MW, Jain C, Jain M, Jarvis ED, Kerpedjiev P, Kirsche M, Kolmogorov M, Korlach J, Kremitzki M, Li H, Maduro VV, Marschall T, McCartney AM, McDaniel J, Miller DE, Mullikin JC, Myers EW, Olson ND, Paten B, Peluso P, Pevzner PA, Porubsky D, Potapova T, Rogaev EI, Rosenfeld JA, Salzberg SL, Schneider VA, Sedlazeck FJ, Shafin K, Shew CJ, Shumate A, Sims Y, Smit AFA, Soto DC, Sović I, Storer JM, Streets A, Sullivan BA, Thibaud-Nissen F, Torrance J, Wagner J, Walenz BP, Wenger A, Wood JMD, Xiao CL, Yan SM, Young AC, Zarate S, Surti U, McCoy RC, Dennis MY, Alexandrov IA, Gerton JL, O'Neill RJ, Timp W, Zook JM, Schatz MC, Eichler EE, Miga KH, Phillippy AM. The complete sequence of a human genome. Science, 2022, 376(6588): 44-53.
doi: 10.1126/science.abj6987 pmid: 35357919 |
| [1] | 陈小舒, 陈家璧. 动物性染色体剂量补偿[J]. 遗传, 2025, 47(2): 200-210. |
| [2] | 刘奇, 施鹏. 脊椎动物回声定位发声和听觉机制研究进展[J]. 遗传, 2025, 47(2): 237-257. |
| [3] | 杨漫宇, 姚方杰, 杨足君, 杨恩年. 六倍体小黑麦×六倍体小麦杂交后代中染色体遗传与结构变异鉴定[J]. 遗传, 2024, 46(1): 63-77. |
| [4] | 马茜, 周少岚, 党洁, 霍正浩, 马占兵. 失活X染色体基因逃逸与系统性红斑狼疮的性别二态性[J]. 遗传, 2024, 46(1): 18-33. |
| [5] | 张翌, 吴志英. 伴皮质下梗死和白质脑病的常染色体显性遗传性脑动脉病的发病机制及治疗研究进展[J]. 遗传, 2023, 45(7): 568-579. |
| [6] | 吴丹丹, 朱明昆, 方忠艳, 马伟. 植物B染色体的分子结构组成及遗传机制研究进展[J]. 遗传, 2022, 44(9): 772-782. |
| [7] | 王飞, 王萌, 张兴华, 宇克莉, 郑连斌, 杨亚军. 中国西南地区3个隔离人群遗传亚结构分析[J]. 遗传, 2022, 44(5): 424-431. |
| [8] | 王海燕, 龚志云, 蒋甲福, 周宝良, 娄群峰, 曹清河, 席梦利, 陈佩度, 顾铭洪, 张天真, 陈发棣, 陈劲枫, 李宗芸, 王秀娥. 江苏省植物细胞遗传学研究回顾与展望[J]. 遗传, 2021, 43(5): 397-424. |
| [9] | 刘志勇, 任贺, 陈冲, 张京晶, 张晓梦, 石妍, 石林玉, 陈滢, 程凤, 贾莉, 陈曼, 范庆炜, 张家榕, 李万婷, 王萌春, 任子林, 刘雅诚, 倪铭, 孙宏钰, 严江伟. 基于有限突变模型和大规模数据的19个常染色体STR的实际突变率研究[J]. 遗传, 2021, 43(10): 949-961. |
| [10] | 史晓黎,何伊琳,凌宏清. 小麦A基因组测序与进化研究进展[J]. 遗传, 2019, 41(9): 836-844. |
| [11] | 王燕超,马晓燕,孙筱放,冼嘉嘉,李少英,何文智,王晓蔓,黎青. Y染色体微缺失人群中Y-STR等位基因缺失模式分析[J]. 遗传, 2019, 41(3): 243-253. |
| [12] | 孟玉,杨若林. 基于基因家族大小的比较研究脊椎动物的适应性进化[J]. 遗传, 2019, 41(2): 158-174. |
| [13] | 彭继苹, 袁海明. 染色体微阵列分析技术在2600例流产物中的应用[J]. 遗传, 2018, 40(9): 779-788. |
| [14] | 杨德卫, 郑向华, 程朝平, 叶宁, 黄凤凰, 叶新福. 基于CSSLs群体定位和图位克隆水稻长芒基因GAD1-2[J]. 遗传, 2018, 40(12): 1101-1111. |
| [15] | 马磊, 张婷婷. 应用嵌合基因实例拓展遗传学染色体畸变的教学[J]. 遗传, 2018, 40(12): 1129-1135. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||

www.chinagene.cn
备案号:京ICP备09063187号-4
总访问:,今日访问:,当前在线:

