遗传 ›› 2025, Vol. 47 ›› Issue (8): 903-927.doi: 10.16288/j.yczz.24-373
雷定伟1( ), 顾睿初2(
), 顾睿初2( ), 谢晓雪2, 丁时之3, 温翰3,4,5(
), 谢晓雪2, 丁时之3, 温翰3,4,5( )
)
收稿日期:2025-02-26
修回日期:2025-06-19
出版日期:2025-06-24
发布日期:2025-06-24
通讯作者:
温翰,博士,副研究员,研究方向:分子动力学模拟,RNA科学,系统生物学。E-mail: wenh@aisi.ac.cn作者简介:雷定伟,本科生,专业方向:药学。E-mail: dwlei@stu.pku.edu.cn雷定伟和顾睿初并列第一作者。
基金资助:
Dingwei Lei1( ), Ruichu Gu2(
), Ruichu Gu2( ), Xiaoxue Xie2, Shizhi Ding3, Han Wen3,4,5(
), Xiaoxue Xie2, Shizhi Ding3, Han Wen3,4,5( )
)
Received:2025-02-26
Revised:2025-06-19
Published:2025-06-24
Online:2025-06-24
Supported by:摘要:
N6-甲基腺嘌呤(m6A)修饰是真核生物mRNA中最丰富的修饰形式,对mRNA的剪接、加工、降解和翻译的调控具有关键作用。本文介绍了计算方法在m6A修饰研究中的应用,主要有数据驱动的方法预测m6A位点以及基于分子动力学方法探究m6A相关生物机制。文章首先回顾了m6A检测技术的发展历程,阐述了相应的数据处理方法,并整理了现有公开数据集,为计算模型的构建奠定数据基础。接着重点讨论机器学习与深度学习模型在m6A位点预测中的研究进展。最后描述了分子动力学模拟在解析m6A相关分子机制的贡献,展示了计算方法如何促进对这一复杂的表观遗传调控过程的理解。通过系统梳理相关内容,本文深入探讨了计算方法在m6A修饰领域的最新研究进展及其应用价值,为m6A相关的深入研究提供新的思路与启示。
雷定伟, 顾睿初, 谢晓雪, 丁时之, 温翰. 计算方法在m6A研究中的应用及展望[J]. 遗传, 2025, 47(8): 903-927.
Dingwei Lei, Ruichu Gu, Xiaoxue Xie, Shizhi Ding, Han Wen. Application and prospects of current computational methods in m6A research: a comprehensive review[J]. Hereditas(Beijing), 2025, 47(8): 903-927.
图1
m6A修饰的动态调控机制及计算生物学在其中的应用概览 上方展示了m6A修饰的“书写”“擦除”和“读取”过程:在“书写”环节,由METTL3、METTL14等组成的甲基转移酶复合体,以S-腺苷甲硫氨酸(S-adenosylmethionine,SAM)为甲基供体,将m6A修饰添加到RNA特定位点;“擦除”环节中,FTO和ALKBH5酶通过催化去甲基化反应去除m6A修饰;“读取”环节里,YTH家族蛋白等识别蛋白通过特异性结合m6A修饰碱基,介导RNA代谢调控。下方呈现了计算生物学的两大应用方向:在数据驱动的修饰位点预测中,整体流程涵盖了实验检测到数据整理收集,算法模型搭建以及修饰位点预测;基于物理模型的机制探究则通过模拟系统构建、参数调优、分子动力学模拟以及结果分析,进而揭示m6A相关分子互作机制。绘制于 https://BioRender.com。"
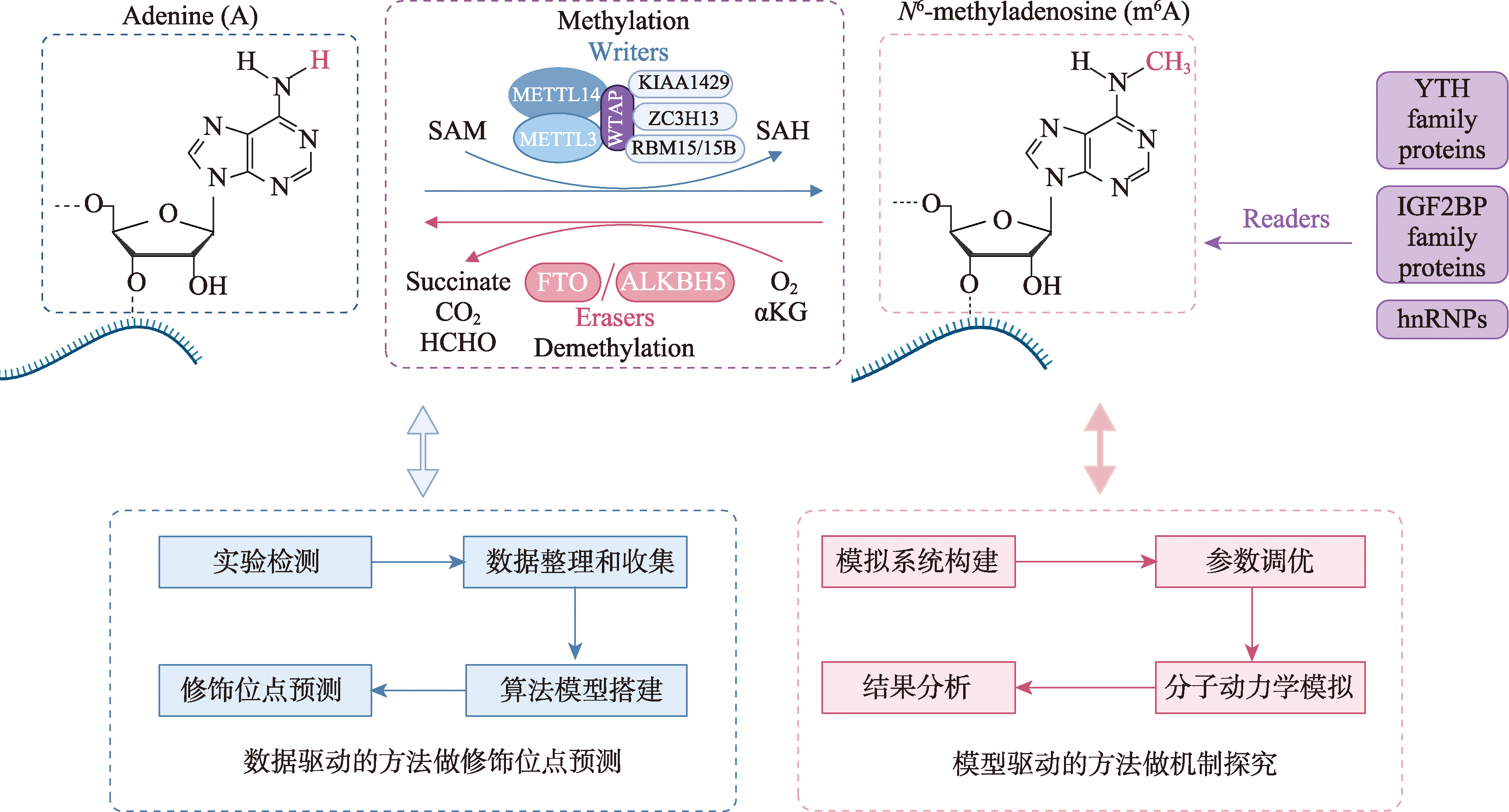
表1
m6A相关的检测方法总结"
| 分类 | 方法 | 分辨率 | RNA 输入量 | 优势 | 局限性 | 发布年份及参考文献 |
|---|---|---|---|---|---|---|
| 抗体依赖的免疫沉淀法 | m6A-seq/ MeRIP-seq | 100~200 nt | 2~400 μg mRNA | 简单的RNA库制备步骤 | 1. 分辨率低; 2. 准确度低; 3. 特异性差,不能区分m6A和m6Am | 2012[ |
| PA-m6A-seq | ~23 nt | 12 μg mRNA | 准确度高; 兼容核酸-蛋白相互作用的研究 | 对样本输入量的需求高; 对培养环境要求高; 交联率低 | 2015[ | |
| m6A-CLIP-seq | ~100 nt | 1 μg mRNA | 1. 对样本输入量要求低; 2. 利用突变和截断特征来识别峰内的m6A位点确保了高水平的特异性; 3. 每个转录本可以检测多个m6A位点 | 1. 缺乏化学计量信息; 2. 无法区分紧密沉积的m6A簇; 3. 不是直接鉴定单m6A位点,而是从相邻嘧啶位点突变中进行推断 | 2018[ | |
| miCLIP-seq | 单核苷酸 | 20 μg mRNA | 1. 特异性和准确度高; 2. 无需对细胞进行修饰核苷酸预处理; 3. 是最被广泛使用的m6A测序方法 | 1. 缺乏化学计量信息; 2. 对样本输入量要求高; 3. RNA库制备流程复杂; 4. 交联率(crosslinking yield)低导致假阳性结果多; 5. 不是直接鉴定单m6A位点,而是从相邻嘧啶位点突变中进行推断 | 2017[ | |
| m6A-LAIC-seq | 100~200 nt | 2 μg 2× poly(A)选择的mRNA | 1. 包含半化学计量信息; 2. 阐明组织或细胞水平下m6A水平的特异性; 3. 准确度和灵敏度高 | 1. 不能区分m6A和m6Am; 2. 实验过程复杂; 3. 需要抗体浓度的经验滴定; 4. 分辨率相对较低 | 2016[ | |
| m6AISH-PLA | 单核苷酸 | − | 1. 可以在mRNA的特定位置识别m6A; 2. 实验条件温和,有助于保存细胞结构和形态; 3. 揭示特定m6A RNA的空间位置 | 1. 通量相对较低; 2. 无法区分短序列范围内的m6A位点 | 2021[ | |
| 特异性酶结合法 | MAZTER-seq | 单核苷酸 | 100 ng mRNA | 1. 可以获取mRNA上ACA位点的m6A化学计量信息; 2. 假阳性概率低; 3. 与抗体依赖的免疫沉淀法相比,实验过程相对简单; 4. 该方法能够区分m6A和m6Am | 1. 检测区域有限(仅检测ACA序列上下文中的m6A位点); 2. 无法区分近距离的ACA位点; 3. 对于绝对量化,切割效率需要在甲基化缺陷背景下进行标准化; 4. MazF并非特异性识别ACA位点,在类似ACA的序列上也观察到少量的切割,如ACG或AAA,导致确度降低 | 2019[ |
| DART-seq | 单核苷酸 | 10 ng~1μg total RNA | 1. 对样本输入量要求低; 2. 能够区分m6A和m6Am | 1. 对低丰度m6A位点的灵敏度低; 2. 可能由于非特异性C-to-U编辑事件而产生假阳性信号 | 2019[ | |
| m6A-REF-seq | 单核苷酸 | 100 ng mRNA | 准确性和可靠性高 | 仅检测ACA序列上下文中的m6A位点 | 2019[ | |
| m6A-SEAL-seq | 100-200 nt | 5 μg mRNA | 1. 对样本输入量要求低; 2. 特异性和可靠性较高; 3. 对于低丰度位点的检测很灵敏 | 1. 分辨率低; 2. 缺乏化学计量信息; 3. 操作步骤多、耗时长 | 2020[ | |
| scDART-seq | 单核苷酸 | 900 ng total RNA | 1. 第一种单细胞水平的m6A检测方法; 2. 准确度高 | 1. 需要在感兴趣的细胞或组织中表达外源基因APOBEC1-YTH; 2. 难以应用于组织样品 | 2022[ | |
| eTAM-seq | 单核苷酸 | 250 ng total RNA | 1. 实现了RNA每个位点的定量化学计量信息; 2. 对样本输入量要求低; 3. 灵敏度高 | 1. 对甲基化水平低的位点不敏感; 2. 需要一个阴性对照文库(由体外转录的转录组制备)和一个专用的统计模型; 3. 需要表达和纯化TadA8.20酶 | 2023[ | |
| 基于化学处理的方法 | SCARLET | 单核苷酸 | 1 μg mRNA | 1. 在特定位点精确测量m6A的化学计量; 2. 对于生物样品中的低丰度转录本准确度高 | 1. 耗时长、步骤复杂; 2. 对样本输入量需求高; 3. 使用放射性试剂 | 2013[ |
| SELECT | 单核苷酸 | >1 μg total RNA | 1. 包含化学计量信息; 2. 准备步骤相对简单 | 1. 假阳性概率高; 2. 通量低; 3. 需要引入对照组并生成标准曲线以完成检测和定量分析 | 2018[ | |
| m6A-label-seq | 单核苷酸 | 50ng total RNA | 1. 准确度高; 2. 适用于细胞内RNA多种不同甲基化序列(m6A motif)的鉴定; 3. 对测定m6A簇修饰有优势 | 1. 缺乏化学计量学信息; 2. 在标记产率和标记时间窗口尺度方面需要优化提高 | 2020[ | |
| m6A-ORL-seq | 单核苷酸 | 30 ng total RNA或5ng mRNA | 1. 准确度和灵敏度高; 2. 测序成本相对较低; 3. 可用于DNA中m6A水平的估计和6mdA的检测 | 在生信分析中易发生数据突变 | 2022[ | |
| m6A-SAC-seq | 单核苷酸 | 2~50 ng mRNA | 1. 对样本输入量要求低; 2. 包含化学计量信息; 3. 准确度高; 4. 可以应用于各种类型的生物样品,包括新鲜和冷冻组织以及福尔马林固定或石蜡包埋的样品; 5. 可以识别大量m6A位点 | MjDim1对GAC基序的明显偏好导致在检测AAC位点方面存在限制 | 2023[ | |
| GLORI-seq | 单核苷酸 | 100 ng | 1. 实现对m6A进行绝对定量评估; 2. 准确度、特异性和灵敏度高; 3. 结果高度可重复; 4. 揭示了 m6A 的定量图谱 | 1. 测序成本相对较高; 2. 无法区分m6A和其他A修饰,例如m1A或m6Am; 3. 灵敏度可能会受到化学标记效率的影响,并且会根据实验设置而变化 | 2023[ | |
| 基于纳米孔直接测序的方法 | Nanopore DRS | 单核苷酸 | − | 1. 不受试剂偏差与扩征偏差的影响; 2. 可以测复杂转录本的全修饰图景 | 1. 修饰引起的电信号偏弱,误差较大; 2.数据分析困难 | 2019[ |
表2
m6A数据处理方法"
| 方法 | 输入文件 | 峰值检测 | 算法 | 优势 | 局限性 | 发布年份及参考文献 | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| MACS | BAM | 支持,主要用于ChIP-seq数据 | 利用泊松分布模型来评估测序深度的富集程度 | 可有效地处理ChIP-Seq数据中的噪声 | 需要依赖合适的参数设置和对照组数据以提高稳定性 | 2008[ | ||||
| exomePeak | BAM | 支持,基于exomePeak | 对所有重复样本的平均标准化计数进行 Fisher 精确检验 | 灵敏度(真阳性率)较高,真发现率(TDR)表现突出 | 1. 使用合并的读取计数,忽略了重复样本间的异质性; 2. 假发现率(FDR)/I型错误控制较差; 3. 大样本时运行时间较长 | 2014[ | ||||
| FET-HMM | Read count matrix | 支持,基于exomePeak | 结合Fisher精确检验和隐马尔可夫模型(HMM)以提高DMR(差异甲基化区)检测的空间分辨率 | 1. 兼具高真发现率(TDR)和灵敏度; 2. I 型错误控制达到理论最优水平 | 1.使用合并的读取计数,忽略了重复样本间的异质性 2. 假发现率(FDR)/假阳性控制较差 | 2015[ | ||||
| MeTDiff | BAM | 支持,基于HEPeak | 针对原始IP计数构建 Beta-二项分布模型,并考虑IP和输入样本的总计数 | 内存消耗少 | 1. 未校正测序深度变异的影响 2. 在小样本量情况下检测效力较差 3. 大样本数据运行时间较长 | 2015[ | ||||
| MeTPeaK | BAM | 支持,主要用于MeRIP-seq数据 | 采用隐马尔可夫模型和Beta分布分层建模 | 1. 隐马尔可夫模型能够捕捉序列数据中的隐含状态; 2. 适用于处理有复杂噪声的数据 | 1. 计算资源需求较高; 2. 依赖高质量数据与精确参数配置 | 2016[ | ||||
| MATK | BAM | 支持,主要用于MeRIP-seq数据 | 用于m6A修饰检测的工具包,集成了包括统计模型、机器学习、深度学习等多种方法 | 1. 可根据数据特点选择最优方法; 2. 可扩展性好 | 不同算法的性能可能因数据集而异,需要进行比较和选择 | 2016* | ||||
| DRME | Read count matrix | 支持,基于exomePeak | 针对原始IP和输入计数构建负二项分布模型,仅使用输入计数估计基线表达 | 即使在小样本和低表达情况下,仍能保持最高灵敏度 | 1. 变异建模不合理; 2. P值要求“宽松”,导致最高的假发现率(FDR)和I型错误 | 2016[ | ||||
| QNB | Read count matrix | 支持,基于 exomePeak | 针对原始IP和输入计数构建负二项分布模型,并同时使用IP和输入计数估计基线表达 | 较低的假发现率(FDR) | 变异建模不合理 | 2017[ | ||||
| exomePeak2 | BAM | 支持,基于 exomePeak2 | 应用DESeq2进行回归分析,并通过三次样条展开的泊松广义线性模型(GLM)调整GC含量偏差 | 1. 高真发现率(TDR)和较低的假发现率(FDR)控制较优; 2. p值分布合理 | 1.不能在模型中纳入额外的实验因素; 2. 内存消耗较大 | 2019[ | ||||
| RADAR | BAM | 否 | 针对预处理的IP计数数据构建泊松随机效应模型,并允许纳入混杂因素 | 首个考虑混杂因素的m6A分析方法 | 1. 对预处理数据的分布假设不合理; 2. 运行时间较长 | 2019[ | ||||
| TRESS | BAM | 支持,基于TRESS | 针对原始IP和输入计数数据构建负二项分布模型,并允许纳入混杂因素 | 1. 真发现率(TDR)高; 2. 假发现率(FDR)/I 型错误控制较优; 3. P值分布合理; 4. 运行时间最短,内存消耗低 | 在小样本条件下灵敏度较低 | 2022[ | ||||
| m6ACali | BAM | 支持,主要用于MeRIP-seq数据 | 基于机器学习方法整合序列特征和基因注释信息 | 减少了假阳性位点 | 需要较多的训练数据和特征信息 | 2024[ | ||||
表3
m6A相关的数据库总结"
| 数据库名称 | 主要技术 | 数据类型 | 覆盖物种 | 位点数量 | 数据来源 | 发布年 份及参 考文献 | |
|---|---|---|---|---|---|---|---|
| MeT-DB | MeRIP-seq | m6A峰 | 人类(Homo sapiens),小鼠(Mus musculus) | 约30万个m6A修饰位点 | 公共数据库 | 2014[ | |
| RMBase | m6A-seq,位点预测 | 单碱基位点的修饰 | 人类,小鼠和酵母(Saccharomyces cerevisiae) | 大约124,200个m6A修饰位点 | 来自GEO数据库及论文的补充材料 | 2015[ | |
| MeT-DB V2.0 | MeRIP-seq | m6A峰,单碱基位点的修饰 | 人类,小鼠等7个物种 | 大约370万个m6A修饰位点 | 26项独立研究 | 2017[ | |
| m6AVar | miCLIP/PA-m6A-seq, MeRIP-seq, 全转录组预测 | m6A相关遗传变异 | 人类 | 16,132个高置信度,71,321个中等置信度,326,915个低置信度m6A修饰位点 | GWAS、ClinVar | 2017[ | |
| RMBase V2.0 | m6A-seq,MeRIP- seq,位点预测 | 单碱基位点的修饰 | 人类,小鼠等13个物种 | 约137万个m6A修饰位点 | 47项研究中的约600个数据集 | 2017[ | |
| CVm6A | MeRIP-seq,m6A-CLIP-seq | m6A峰 | 人类,小鼠 | 40,950个人类细胞系的m6A修饰位点,179,201个鼠类细胞系的m6A修饰位点 | 公共数据库 | 2019[ | |
| M6A2Target, RM2Target | 低通量和高通量研究 | WERs靶标 | 人类和小鼠 | − | NCBI、PubMed、GEO和SRA等数据库 | 2020[ | |
| M6ADD | 人工筛选和高通量测序 | m6A-疾病关联 | 人类 | 409,229个m6A-疾病关联 | 实验数据、GWAS、ClinVar | 2020[ | |
| m6A-Atlas | 7种单碱基分辨率技术 | 单碱基分辨率m6A位点 | 人类、小鼠和黑猩猩(Pan troglodytes)等7个物种 | 442,162个m6A修饰位点 | 高通量测序数据 | 2020[ | |
| RMVar | − | m6A修饰相关遗传变异 | 人类 | m6A修饰的1,461,691个遗传变异 | 实验数据、GWAS、ClinVar | 2020[ | |
| REPIC | MeRIP-seq,m6A-seq | m6A峰 | 11种生物 | 大约1,000万个m6A峰 | 49项研究的672个样本 | 2020[ | |
| m6A-TSHub | − | m6A峰 | 人类 | 来自23个人类组织的184,554个修饰位点和499,369个来自25种肿瘤的修饰位点 | GEO,NGDC | 2022[ | |
| DirectRMDB | 直接RNA测序(DRS) | 单碱基分辨率m6A位点 | 人类、小鼠等25个物种 | 586,167个修饰数据 | 来源于39项独立研究 | 2022[ | |
| PRMD | MeRIP-seq | m6A峰 | 20种植物 | 6,816,278个m6A峰 | 主要来自EnsemblPlants | 2023[ | |
| RMBase V3.0 | 单碱基分辨率以及有限分辨率的测序技术 | m6A峰或单碱基位点的修饰 | 与m6A修饰相关的主要有人类,小鼠等物种 | 67万个修饰数据 | GEO | 2023[ | |
| RMVar 2.0 | 高通量测序,单碱基分辨率的修饰位点分析等 | m6A峰或单碱基位点的修饰 | 人、小鼠 | 1,680,598个m6A修饰数据 | 文献报道,公共数据库 | 2024[ | |
| m6A-Atlas V2.0 | 13种单碱基分辨率技术,2种单细胞m6A分析技术,MeRIP-seq | m6A峰,单碱基位点的修饰 | 多物种 | 797,091个m6A单碱基修饰位点 | 公共数据库,文献报道 | 2024[ | |
表4
m6A修饰位点预测模型总结"
| 模型名称 | 物种 | 模型架构 | 特点 | 数据来源 | 发布年份及参考文献 |
|---|---|---|---|---|---|
| DeepM6ASeq | 人类、小鼠、斑马鱼(Danio rerio) | CNN+BiLSTM | 提取序列特征,显著性图可视化,预测性能优于传统分类器,发现新m6A阅读器FMR1 | SRAMP[ | 2018[ |
| TDm6A | 人类 | CNN+LSTM+迁移学习 | 学习常见和细胞类型特异性motif,预测lncRNA m6A位点,适应不同细胞类型 | SRAMP[ | 2020[ |
| MASS | 人类、小鼠、恒河猴(Macaca mulatta)、大鼠、猪和斑马鱼 | BiLSTM+CNN+Multi- Head Attention | 捕捉多物种共享特征,课程学习策略,减少物种偏差,增强泛化能力和解释性 | RMBase v2.0[ | 2021[ |
| Deepm6A-MT | 人类、大鼠(Rattus norvegicus)和小鼠 | Bi-GRU+CNN | 专注多组织的m6A预测,融合局部和全局信息,多种序列表示方法,提升预测精度,用户友好网络服务器 | iRNA-m6A[ | 2024[ |
| deepSRAMP | 人类、小鼠、大鼠 | Transformer+RNN | 跨物种泛化能力强,识别isoform水平m6A修饰位点,引入注意力机制,处理复杂多转录本基因 | m6A-Atlas v2.0[ | 2024[ |
| m6ATM | 人类 | WaveNet+Dual-Stream Multiple Instance Learning (DSMIL) | 结合1D卷积和MIL策略提取m6A特征,使用纳米孔测序数据进行单碱基分辨率m6A预测,支持m6A化学计量估计 | GSE124309、 GSE132971、GSE265754、 GSE265867和PRJEB40872 | 2024[ |
| pum6a | 人类、小鼠 | Attention-based Positive and Unlabeled Multi-Instance Learning (MIL) | 结合电信号特征和碱基比对数据,使用加权Noisy-OR概率机制,增强低覆盖位点的m6A检测灵敏度和准确性 | HEK293T[ | 2025[ |
| TandemMod | 人类、水稻(Oryza sativa) | 1D-CNN+BiLSTM+ Attention | 采用迁移学习,提高新修饰类型的检测效率;可处理单细胞或批量数据,检测修饰比例 | IVET[ | 2024[ |
| [1] |
Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell, 2017, 169(7): 1187-1200.
pmid: 28622506 |
| [2] | Yang Y, Chen YS, Sun BF, Yang YG. RNA methylation: regulations and mechanisms. Hereditas (Beijing), 2018, 40(11): 964-976. |
| 杨莹, 陈宇晟, 孙宝发, 杨运桂. RNA甲基化修饰调控和规律. 遗传, 2018, 40(11): 964-976. | |
| [3] |
Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci USA, 1974, 71(10): 3971-3975.
pmid: 4372599 |
| [4] |
Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′UTRs and near stop codons. Cell, 2012, 149(7): 1635-1646.
pmid: 22608085 |
| [5] |
Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, Sorek R, Rechavi G. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature, 2012, 485(7397): 201-206.
pmid: 22575960 |
| [6] |
Ke SD, Alemu EA, Mertens C, Gantman EC, Fak JJ, Mele A, Haripal B, Zucker-Scharff I, Moore MJ, Park CY, Vågbø CB, Kusśnierczyk A, Klungland A, Darnell JE JR, Darnell RB. A majority of m6A residues are in the last exons, allowing the potential for 3' UTR regulation. Genes Dev, 2015, 29(19): 2037-2053.
pmid: 26404942 |
| [7] |
Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet, 2014, 15(5): 293-306.
pmid: 24662220 |
| [8] |
Liu WW, Zheng SQ, Li T, Fei YF, Wang C, Zhang S, Wang F, Jiang GM, Wang H. RNA modifications in cellular metabolism: implications for metabolism- targeted therapy and immunotherapy. Signal Transduct Target Ther, 2024, 9(1): 70.
pmid: 38531882 |
| [9] |
Lee M, Kim B, Kim VN. Emerging roles of RNA modification: m6A and U-tail. Cell, 2014, 158(5): 980-987.
pmid: 25171402 |
| [10] | Zhang X, Jia GF. RNA epigenetic modification: N6-methyladenosine. Hereditas (Beijing), 2016, 38(4): 275-288. |
| 张笑, 贾桂芳. RNA表观遗传修饰:N6-甲基腺嘌呤. 遗传, 2016, 38(4): 275-288. | |
| [11] |
Liu JZ, Yue YN, Han DL, Wang X, Fu Y, Zhang L, Jia GF, Yu M, Lu ZK, Deng X, Dai Q, Chen WZ, He C. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol, 2014, 10(2): 93-95.
pmid: 24316715 |
| [12] |
Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, Zhao X, Li A, Yang Y, Dahal U, Lou XM, Liu X, Huang J, Yuan WP, Zhu XF, Cheng T, Zhao YL, Wang X, Danielsen JMR, Liu F, Yang YG. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res, 2014, 24(2): 177-189.
pmid: 24407421 |
| [13] |
Yue YN, Liu J, Cui XL, Cao J, Luo GZ, Zhang ZZ, Cheng T, Gao MS, Shu X, Ma HH, Wang FQ, Wang XX, Shen B, Wang YZ, Feng XH, He C, Liu JZ. VIRMA mediates preferential m6A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation. Cell Discov, 2018, 4: 10.
pmid: 29507755 |
| [14] |
Wen J, Lv RT, Ma HH, Shen HJ, He CX, Wang JH, Jiao FF, Liu H, Yang PY, Tan L, Lan F, Shi YJG, He C, Shi Y, Diao JB. Zc3h13 Regulates nuclear RNA m6A methylation and mouse embryonic stem cell self-renewal. Mol Cell, 2018, 69(6): 1028-1038.e6.
pmid: 29547716 |
| [15] |
Wang H. The RNA m6A writer RBM15 contributes to the progression of esophageal squamous cell carcinoma by regulating miR-3605-5p/KRT4 pathway. Heliyon, 2024, 10(2): e24459.
pmid: 38312624 |
| [16] |
Azzam SK, Alsafar H, Sajini AA. FTO m6A demethylase in obesity and cancer: implications and underlying molecular mechanisms. Int J Mol Sci, 2022, 23(7): 3800.
pmid: 35409166 |
| [17] |
Zheng GQ, Dahl John A, Niu YM, Fedorcsak P, Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH, Lu ZK, Bosmans RPG, Dai Q, Hao YJ, Yang X, Zhao WM, Tong WM, Wang XJ, Bogdan F, Furu K, Fu Y, Jia GF, Zhao X, Liu J, Krokan HE, Klungland A, Yang YG, He C. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Molecular Cell, 2013, 49(1): 18-29.
pmid: 23177736 |
| [18] |
Chen L, Gao Y, Xu SM, Yuan JX, Wang M, Li TY, Gong J. N6-methyladenosine reader YTHDF family in biological processes: Structures, roles, and mechanisms. Front Immunol, 2023, 14: 1162607.
pmid: 36999016 |
| [19] |
Wang X, Lu ZK, Gomez A, Hon GC, Yue YN, Han DL, Fu Y, Parisien M, Dai Q, Jia GF, Ren B, Pan T, He C. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature, 2014, 505(7481): 117-120.
pmid: 24284625 |
| [20] | Song PH, Ma LJ, Yan D. Exon junction complex modulates the formation of the m6A epitranscriptome. Hereditas (Beijing), 2023, 45(6): 464-471. |
| 宋鹏辉, 马丽娟, 严冬. 外显子拼接复合体塑造m6A表观转录组的形成. 遗传, 2023, 45(6): 464-471. | |
| [21] |
Pan YT, Ma P, Liu Y, Li W, Shu YQ. Multiple functions of m6A RNA methylation in cancer. J Hematol Oncol, 2018, 11(1): 48.
pmid: 29587823 |
| [22] |
Jiang XL, Liu BY, Nie Z, Duan LC, Xiong QX, Jin ZX, Yang CP, Chen YB. The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther, 2021, 6(1): 74.
pmid: 33611339 |
| [23] | Wang TG, Ye M. Advances on the roles of m6A in tumorigenesis. Hereditas (Beijing), 2018, 40(12): 1055-1065. |
| 王天工, 叶孟. m6A甲基化与肿瘤研究进展. 遗传, 2018, 40(12): 1055-1065. | |
| [24] |
Chen K, Lu ZK, Wang X, Fu Y, Luo GZ, Liu N, Han DL, Dominissini D, Dai Q, Pan T, He C. High-resolution N6-methyladenosine (m6A) map using photo- crosslinking-assisted m6A sequencing. Angew Chem Int Ed Engl, 2015, 54(5): 1587-1590.
pmid: 25491922 |
| [25] |
Hsu PJ, He C. High-resolution mapping of N6-Methyladenosine using m6A crosslinking immunoprecipitation sequencing (m6A-CLIP-Seq). Methods Mol Biol, 2019, 1870: 69-79.
pmid: 30539547 |
| [26] |
Grozhik AV, Linder B, Olarerin-George AO, Jaffrey SR. Mapping m6A at individual-nucleotide resolution using crosslinking and immunoprecipitation (miCLIP). Methods Mol Biol, 2017, 1562: 55-78.
pmid: 28349454 |
| [27] |
Molinie B, Wang JK, Lim KS, Hillebrand R, Lu ZX, Van Wittenberghe N, Howard BD, Daneshvar K, Mullen AC, Dedon P. m6A-LAIC-seq reveals the census and complexity of the m6A epitranscriptome. Nat Methods, 2016, 13(8): 692-698.
pmid: 27376769 |
| [28] | Ren XJ, Deng RJ, Zhang KX, Sun YP, Li Y, Li JH. Single-cell imaging of m6A modified RNA using m6A-specific in situ hybridization mediated proximity ligation assay (m6AISH-PLA). Angew Chem Int Edit, 2021, 60(42): 22646-22651. |
| [29] |
Garcia-Campos MA, Edelheit S, Toth U, Safra M, Shachar R, Viukov S, Winkler R, Nir R, Lasman L, Brandis A. Deciphering the “m6A code” via antibody- independent quantitative profiling. Cell, 2019, 178(3): 731-747.e16.
pmid: 31257032 |
| [30] |
Meyer KD. DART-seq: an antibody-free method for global m6A detection. Nat Methods, 2019, 16(12): 1275-1280.
pmid: 31548708 |
| [31] |
Zhang Z, Chen LQ, Zhao YL, Yang CG, Roundtree IA, Zhang ZJ, Ren J, Xie W, He C, Luo GZ. Single-base mapping of m6A by an antibody-independent method. Sci Adv, 2019, 5(7): eaax0250.
pmid: 31281898 |
| [32] |
Wang Y, Xiao Y, Dong SQ, Yu Q, Jia GF. Antibody-free enzyme-assisted chemical approach for detection of N6-methyladenosine. Nat Chem Biol, 2020, 16(8): 896-903.
pmid: 32341502 |
| [33] |
Tegowski M, Flamand MN, Meyer KD. scDART-seq reveals distinct m6A signatures and mRNA methylation heterogeneity in single cells. Mol Cell, 2022, 82(4): 868-878.e10.
pmid: 35081365 |
| [34] |
Xiao YL, Liu S, Ge RQ, Wu Y, He C, Chen MJ, Tang WX. Transcriptome-wide profiling and quantification of N6-methyladenosine by enzyme-assisted adenosine deamination. Nat Biotechnol, 2023, 41(7): 993-1003.
pmid: 36593412 |
| [35] |
Liu N, Parisien M, Dai Q, Zheng GQ, He C, Pan T. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA, 2013, 19(12): 1848-1856.
pmid: 24141618 |
| [36] |
Xiao Y, Wang Y, Tang Q, Wei LH, Zhang X, Jia G. An elongation-and ligation-based qPCR amplification method for the radiolabeling-free detection of locus- specific N6-methyladenosine modification. Angewe Chem Int Ed Engl, 2018, 57(49): 15995-16000.
pmid: 30345651 |
| [37] |
Shu X, Cao J, Cheng MH, Xiang SY, Gao MS, Li T, Ying XR, Wang FQ, Yue YN, Lu Z, Dai Q, Cui XL, Ma LJ, Wang YZ, He C, Feng XH, Liu JZ. A metabolic labeling method detects m6A transcriptome-wide at single base resolution. Nat Chem Biol, 2020, 16(8): 887-895.
pmid: 32341503 |
| [38] |
Xie YL, Han SQ, Li QM, Fang ZT, Yang W, Wei Q, Wang YF, Zhou Y, Weng X, Zhou X. Transcriptome-wide profiling of N6-methyladenosine via a selective chemical labeling method. Chem Sci, 2022, 13(41): 12149-12157.
pmid: 36349098 |
| [39] |
Ge RQ, Ye C, Peng Y, Dai Q, Zhao YT, Liu S, Wang PL, Hu LL, He C. m6A-SAC-seq for quantitative whole transcriptome m6A profiling. Nat Protoc, 2023, 18(2): 626-657.
pmid: 36434097 |
| [40] |
Liu C, Sun HX, Yi YP, Shen WG, Li K, Xiao Y, Li F, Li YC, Hou Y, Lu B, Liu WQ, Meng HW, Peng JY, Yi CQ, Wang J. Absolute quantification of single-base m6A methylation in the mammalian transcriptome using GLORI. Nat Biotechnol, 2023, 41(3): 355-366.
pmid: 36302990 |
| [41] |
Workman RE, Tang AD, Tang PS, Jain M, Tyson JR, Razaghi R, Zuzarte PC, Gilpatrick T, Payne A, Quick J, Sadowski N, Holmes N, De Jesus JG, Jones KL, Soulette CM, Snutch TP, Loman N, Paten B, Loose M, Simpson JT, Olsen HE, Brooks AN, Akeson M, Timp W. Nanopore native RNA sequencing of a human poly(A) transcriptome. Nat Methods, 2019, 16(12): 1297-1305.
pmid: 31740818 |
| [42] |
Sun HX, Lu B, Zhang ZY, Xiao Y, Zhou Z, Xi L, Li ZC, Jiang Z, Zhang JY, Wang M, Liu C, Ma YC, Peng JY, Wang XJ, Yi CQ. Mild and ultrafast GLORI enables absolute quantification of m6A methylome from low-input samples. Nat Methods, 2025, doi: 10.1038/s41592-025-02680-9.
pmid: 40325216 |
| [43] |
Wei JB, Liu FG, Lu ZK, Fei QL, Ai YX, He PC, Shi HL, Cui XL, Su R, Klungland A, Jia GF, Chen JJ, He C. Differential m6A, m6Am, and m1A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol Cell, 2018, 71(6): 973-985 e975.
pmid: 30197295 |
| [44] |
Garalde D R, Snell E A, Jachimowicz D, Sipos B, Lloyd J H, Bruce M, Pantic N, Admassu T, James P, Warland A, Jordan M, Ciccone J, Serra S, Keenan J, Martin S, Mcneill L, Wallace E J, Jayasinghe L, Wright C, Blasco J, Young S, Brocklebank D, Juul S, Clarke J, Heron A J, Turner DJ. Highly parallel direct RNA sequencing on an array of nanopores. Nat Methods, 2018, 15(3): 201-206.
pmid: 29334379 |
| [45] |
Lorenz DA, Sathe S, Einstein JM, Yeo GW. Direct RNA sequencing enables m6A detection in endogenous transcript isoforms at base-specific resolution. RNA, 2020, 26(1): 19-28.
pmid: 31624092 |
| [46] |
Leger A, Amaral PP, Pandolfini L, Capitanchik C, Capraro F, Miano V, Migliori V, Toolan-Kerr P, Sideri T, Enright AJ, Tzelepis K, van Werven FJ, Luscombe NM, Barbieri I, Ule J, Fitzgerald T, Birney E, Leonardi T, Kouzarides T. RNA modifications detection by comparative nanopore direct RNA sequencing. Nat Commun, 2021, 12(1): 7198.
pmid: 34893601 |
| [47] | Mitchell TM. Machine Learning. New York: McGraw-hill Education, 1997. |
| [48] |
LeCun Y, Bengio Y, Hinton G. Deep learning. Nature, 2015, 521(7553): 436-444.
pmid: 26017442 |
| [49] |
Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-seq (MACS). Genome Biol, 2008, 9(9): R137.
pmid: 18798982 |
| [50] |
Meng J, Cui XD, Rao MK, Chen YD, Huang YF. Exome-based analysis for RNA epigenome sequencing data. Bioinformatics, 2013, 29(12): 1565-1567.
pmid: 23589649 |
| [51] |
Zhang YC, Zhang SW, Liu L, Liu H, Zhang L, Cui XD, Huang YF, Meng J. Spatially enhanced differential RNA methylation analysis from affinity-based sequencing data with hidden markov model. Biomed Res Int, 2015, 2015.
pmid: 26301253 |
| [52] |
Cui XD, Zhang L, Meng J, Rao MK, Chen YD, Huang YF. MeTDiff: a novel differential RNA methylation analysis for MeRIP-Seq data. IEEE/AAM Trans Comput Biol Bioinform, 2018, 15(2): 526-534.
pmid: 29610101 |
| [53] |
Cui X, Meng J, Zhang S, Chen Y, Huang Y. A novel algorithm for calling mRNA m6A peaks by modeling biological variances in MeRIP-seq data. Bioinformatics, 2016, 32(12): i378-i385.
pmid: 27307641 |
| [54] |
Liu L, Zhang SW, Gao F, Zhang YX, Huang YF, Chen RS, Meng J. DRME: count-based differential RNA methylation analysis at small sample size scenario. Anal Biochem, 2016, 499: 15-23.
pmid: 26851340 |
| [55] |
Liu L, Zhang SW, Huang YF, Meng J. QNB: differential RNA methylation analysis for count-based small-sample sequencing data with a quad-negative binomial model. BMC Bioinformatics, 2017, 18(1): 387.
pmid: 28859631 |
| [56] |
Tang Y, Chen K, Song B, Ma J, Wu X, Xu Q, Wei Z, Su J, Liu G, Rong R. m6A-Atlas: a comprehensive knowledgebase for unraveling the N6-methyladenosine (m6A) epitranscriptome. Nucleic Acids Res, 2021, 49(D1): D134-D143.
pmid: 32821938 |
| [57] |
Zhang ZJ, Zhan Q, Eckert M, Zhu A, Chryplewicz A, De Jesus DF, Ren DC, Kulkarni RN, Lengyel E, He C, Chen MJ. RADAR: differential analysis of MeRIP-seq data with a random effect model. Genome Biol, 2019, 20(1): 294.
pmid: 31870409 |
| [58] |
Guo ZX, Shafik AM, Jin P, Wu H. Differential RNA methylation analysis for MeRIP-seq data under general experimental design. Bioinformatics, 2022, 38(20): 4705-4712.
pmid: 36063045 |
| [59] |
Ye HK, Li TL, Rigden DJ, Wei Z. m6ACali: machine learning-powered calibration for accurate m6A detection in MeRIP-Seq. Nucleic Acids Res, 2024, 52(9): 4830-4842.
pmid: 38634812 |
| [60] |
Liu H, Flores MA, Meng J, Zhang L, Zhao XY, Rao MK, Chen YD, Huang YF. MeT-DB: a database of transcriptome methylation in mammalian cells. Nucleic Acids Res, 2015, 43(Database issue): D197-D203.
pmid: 25378335 |
| [61] |
Sun WJ, Li JH, Liu S, Wu J, Zhou H, Qu LH, Yang JH. RMBase: a resource for decoding the landscape of RNA modifications from high-throughput sequencing data. Nucleic Acids Res, 2016, 44(D1): D259-D265.
pmid: 26464443 |
| [62] |
Liu H, Wang HZ, Wei Z, Zhang SY, Hua G, Zhang SW, Zhang L, Gao SJ, Meng J, Chen X, Huang YF. MeT-DB V2. 0: elucidating context-specific functions of N6-methyl- adenosine methyltranscriptome. Nucleic Acids Res, 2018, 46(D1): D281-D287.
pmid: 29126312 |
| [63] |
Zheng YY, Nie P, Peng D, He ZH, Liu MN, Xie YB, Miao YY, Zuo ZX, Ren J. m6AVar: a database of functional variants involved in m6A modification. Nucleic Acids Res, 2018, 46( D1): D139-D145.
pmid: 29036329 |
| [64] |
Xuan JJ, Sun WJ, Lin PH, Zhou KR, Liu S, Zhang LL, Qu LH, Yang JH. RMBase v2.0: deciphering the map of RNA modifications from epitranscriptome sequencing data. Nucleic Acids Res, 2018, 46(D1): D327-D334.
pmid: 29040692 |
| [65] |
Han YJ, Feng J, Xia LJ, Dong X, Zhang XY, Zhang SH, Miao YQ, Xu QD, Xiao S, Zuo ZX, Xia LX, He CJ. CVm6A: a visualization and exploration database for m6As in cell lines. Cells, 2019, 8(2): 168.
pmid: 30781586 |
| [66] |
Deng S, Zhang HW, Zhu KY, Li XY, Ye Y, Li R, Liu XF, Lin DX, Zuo ZX, Zheng J. M6A2Target: a comprehensive database for targets of m6A writers, erasers and readers. Brief Bioinform, 2021, 22(3): bbaa055.
pmid: 32392583 |
| [67] |
Bao XQ, Zhang Y, Li HQ, Teng YY, Ma LX, Chen ZH, Luo XT, Zheng J, Zhao A, Ren J, Zuo ZX. RM2Target:a comprehensive database for targets of writers, erasers and readers of RNA modifications. Nucleic Acids Res, 2023, 51(D1): D269-D279.
pmid: 36300630 |
| [68] |
Zhou DS, Wang HL, Bi FQ, Xing J, Gu Y, Wang C, Zhang MY, Huang Y, Zeng JQ, Wu Q, Zhang Y. M6ADD: a comprehensive database of m6A modifications in diseases. RNA Biol, 2021, 18(12): 2354-2362.
pmid: 33906563 |
| [69] |
Luo XT, Li HQ, Liang JQ, Zhao Q, Xie YB, Ren J, Zuo ZX. RMVar: an updated database of functional variants involved in RNA modifications. Nucleic Acids Res, 2021, 49(D1): D1405-D1412.
pmid: 33021671 |
| [70] |
Liu S, Zhu A, He C, Chen MJ. REPIC:a database for exploring the N6-methyladenosine methylome. Genome Biol, 2020, 21(1): 100.
pmid: 32345346 |
| [71] |
Song BW, Huang DY, Zhang YX, Wei Z, Su JL, De Magalhães JP, Rigden DJ, Meng J, Chen KQ. m6A-TSHub: unveiling the context-specific m6A methylation and m6A-affecting mutations in 23 human tissues. Genomics Proteomics Bioinformatics, 2023, 21(4): 678-694.
pmid: 36096444 |
| [72] |
Zhang YX, Jiang J, Ma JM, Wei Z, Wang Y, Song BW, Meng J, Jia GF, De Magalhães JP, Rigden DJ, Hang DY, Chen KQ. DirectRMDB: a database of post- transcriptional RNA modifications unveiled from direct RNA sequencing technology. Nucleic Acids Res, 2023, 51(D1): D106-D116.
pmid: 36382409 |
| [73] |
Lang XQ, Yu CY, Shen MY, Gu L, Qian Q, Zhou DG, Tan JT, Li YL, Peng X, Diao S, Deng ZJ, Ruan ZH, Xu Z, Xing JL, Li C, Wang RF, Ding CJ, Cao Y, Liu Q. PRMD: an integrated database for plant RNA modifications. Nucleic Acids Res, 2024, 52(D1): D1597-D1613.
pmid: 37831097 |
| [74] |
Xuan JJ, Chen LF, Chen ZR, Pang JJ, Huang JH, Lin JR, Zheng LL, Li B, Qu LH, Yang JH. RMBase v3.0: decode the landscape, mechanisms and functions of RNA modifications. Nucleic Acids Res, 2024, 52(D1): D273-D284.
pmid: 37956310 |
| [75] |
Huang YT, Zhang LWY, Mu WP, Zheng MH, Bao XQ, Li HQ, Luo XT, Ren J, Zuo ZX. RMVar 2.0:an updated database of functional variants in RNA modifications. Nucleic Acids Res, 2024, 53(D1): D275-D283.
pmid: 39436017 |
| [76] |
Liang ZM, Ye HK, Ma JM, Wei Z, Wang Y, Zhang YX, Huang DY, Song BW, Meng J, Rigden DJ, Chen KQ. m6A-Atlas v2.0: updated resources for unraveling the N6-methyladenosine (m6A) epitranscriptome among multiple species. Nucleic Acids Res, 2024, 52(D1): D194-D202.
pmid: 37587690 |
| [77] |
Zhou Y, Zeng P, Li YH, Zhang ZD, Cui QH. SRAMP: prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res, 2016, 44(10): e91.
pmid: 26896799 |
| [78] |
Zhang YQ, Hamada M. DeepM6ASeq: prediction and characterization of m6A-containing sequences using deep learning. BMC Bioinformatics, 2018, 19(Suppl 19): 524.
pmid: 30598068 |
| [79] |
Wang J, Wang LJ. Deep analysis of RNA N6-adenosine methylation (m6A) patterns in human cells. NAR Genom Bioinform, 2020, 2(1): lqaa007.
pmid: 33575554 |
| [80] |
Parker MT, Knop K, Sherwood AV, Schurch NJ, Mackinnon K, Gould PD, Hall AJ, Barton GJ, Simpson GG. Nanopore direct RNA sequencing maps the complexity of Arabidopsis mRNA processing and m6A modification. eLife, 2020, 9: e49658.
pmid: 31931956 |
| [81] |
Xiong YP, He X, Zhao D, Tian TZ, Hong LX, Jiang T, Zeng JY. Modeling multi-species RNA modification through multi-task curriculum learning. Nucleic Acids Res, 2021, 49(7): 3719-3734.
pmid: 33744973 |
| [82] |
Dao FY, Lv H, Yang YH, Zulfiqar H, Gao H, Lin H. Computational identification of N6-methyladenosine sites in multiple tissues of mammals. Comput Struct Biotechnol J, 2020, 18: 1084-1091.
pmid: 32435427 |
| [83] |
Huang GH, Huang XH, Jiang JY. Deepm6A-MT: a deep learning-based method for identifying RNA N6-methyladenosine sites in multiple tissues. Methods, 2024, 226: 1-8.
pmid: 38485031 |
| [84] |
Fan R, Cui CM, Kang BM, Chang ZC, Wang GQ, Cui QH. A combined deep learning framework for mammalian m6A site prediction. Cell Genom, 2024, 4(12): 100697.
pmid: 39571573 |
| [85] |
Yu B, Nagae G, Midorikawa Y, Tatsuno K, Dasgupta B, Aburatani H, Ueda H. m6ATM: a deep learning framework for demystifying the m6A epitranscriptome with Nanopore long-read RNA-seq data. Brief Bioinform, 2024, 25(6): bbae529.
pmid: 39438075 |
| [86] |
Chen Y, Davidson NM, Wan YK, Yao F, Su Y, Gamaarachchi H, Sim A, Patel H, Low HM, Hendra C, Wratten L, Hakkaart C, Sawyer C, Iakovleva V, Lee PL, Xin LX, Ng HEV, Loo JM, Ong XW, Ng HQA, Wang JX, Koh WQC, Poon SYP, Stanojevic D, Tran HD, Lim KHE, Toh SY, Ewels PA, Ng HH, Iyer NG, Thiery A, Chng WJ, Chen LL, Dasgupta R, Sikic M, Chan YS, Tan BOP, Wan Y, Tam WL, Yu Q, Khor CC, Wüstefeld T, Lezhava A, Pratanwanich PN, Love MI, Goh WSS, Ng SB, Oshlack A, SG-NEx consortium, Göke J. A systematic benchmark of Nanopore long-read RNA sequencing for transcript- level analysis in human cell lines. Nat Methods, 2025, 22(4): 801-812.
pmid: 40082608 |
| [87] |
Liu HL, Begik O, Lucas MC, Ramirez JM, Mason CE, Wiener D, Schwartz S, Mattick JS, Smith MA, Novoa EM. Accurate detection of m6A RNA modifications in native RNA sequences. Nat Commun, 2019, 10(1): 4079.
pmid: 31501426 |
| [88] |
Liu CW, Liang H, Wan AH, Xiao M, Sun L, Yu YL, Yan SJ, Deng Y, Liu RN, Fang J, Wang Z, He WL, Wan GH. Decoding the m6A epitranscriptomic landscape for biotechnological applications using a direct RNA sequencing approach. Nat Commun, 2025, 16(1): 798.
pmid: 39824841 |
| [89] |
Wu Y, Shao WN, Yan MX, Wang YQ, Xu PF, Huang GQ, Li XF, Gregory BD, Yang J, Wang HX, Yu X. Transfer learning enables identification of multiple types of RNA modifications using nanopore direct RNA sequencing. Nat Commun, 2024, 15(1): 4049.
pmid: 38744925 |
| [90] |
Chen K, Wei Z, Zhang Q, Wu X, Rong R, Lu Z, Su J, De Magalhães JP, Rigden DJ, Meng J. Whistle: a high-accuracy map of the human N6-methyladenosine (m6A) epitranscriptome predicted using a machine learning approach. Nucleic Acids Res, 2019, 47(7): e41.
pmid: 30993345 |
| [91] |
Chen Z, Zhao P, Li F, Wang Y, Smith AI, Webb GI, Akutsu T, Baggag A, Bensmail H, Song JN. Comprehensive review and assessment of computational methods for predicting RNA post-transcriptional modification sites from RNA sequences. Brief Bioinform, 2020, 21(5): 1676-1696.
pmid: 31714956 |
| [92] |
Maestri S, Furlan M, Mulroney L, Coscujuela Tarrero L, Ugolini C, Dalla Pozza F, Leonardi T, Birney E, Nicassio F, Pelizzola M. Benchmarking of computational methods for m6A profiling with Nanopore direct RNA sequencing. Brief Bioinform, 2024, 25(2): bbae001.
pmid: 38279646 |
| [93] | Huggins DJ, Biggin PC, Dämgen MA, Essex JW, Harris SA, Henchman RH, Khalid S, Kuzmanic A, Laughton CA, Michel J, Mulholland AJ, Rosta E, Sansom MSP. van der Kamp MW. Biomolecular simulations: from dynamics and mechanisms to computational assays of biological activity. Wiley Interdiscip Rev: Comput Mol Sci, 2019, 9(3): e1393. |
| [94] |
Su H, Wang YT, Li HJ. RNA m6A methylation regulators multi-omics analysis in prostate cancer. Front Genet, 2021, 12: 768041.
pmid: 34899855 |
| [95] |
Zhang M, Cai RQ, Liu JJ, Wang YL, He S, Wang Q, Song XF, Wu J, Zhao J. Multi-omics integration analysis reveals the role of N6-methyladenosine in lncRNA translation during glioma stem cell differentiation. Brief Funct Genomics, 2024, 23(6): 806-815.
pmid: 39377261 |
| [96] |
Wu XC, Bai ZP. Multi-omics analysis of m6A modification-related patterns based on m6A regulators and tumor microenvironment infiltration in lung adenocarcinoma. Sci Rep, 2021, 11(1): 20921.
pmid: 34686691 |
| [97] |
Li S, Zhang Y, Liu GY, Song N, Ruan Z, Guo RJ, Tang YL, Cao XQ, Huang XX, Gao T, Hao SJ, Wang QQ, Chang T. Exploring the roles of m6A-modified circRNAs in myasthenia gravis based on multi-omics analysis. Mol Neurobiol, 2025, 62(2): 1694-1704.
pmid: 39017976 |
| [98] |
Liufu C, Luo LX, Pang T, Zheng HH, Yang L, Lu L, Chang S. Integration of multi-omics summary data reveals the role of N6-methyladenosine in neuropsychiatric disorders. Mol Psychiatry, 2024, 29(10): 3141-3450.
pmid: 38684796 |
| [99] |
Karplus M, McCammon JA. Molecular dynamics simulations of biomolecules. Nat Struct Biol, 2002, 9(9): 646-652.
pmid: 12198485 |
| [100] | Rapaport DC. The Art of Molecular Dynamics Simulation. Cambridge University Press, 2004. |
| [101] |
Li YZ, Bedi RK, Wiedmer L, Huang DZ, Śledź P, Caflisch A. Flexible binding of m6A reader protein YTHDC1 to its preferred RNA motif. J Chem Theory Comput, 2019, 15(12): 7004-7014.
pmid: 31670957 |
| [102] |
Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, Wang X, Ma HL, Huang CM, Yang Y, Huang N, Jiang GB, Wang HL, Zhou Q, Wang XJ, Zhao YL, Yang YG. Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol Cell, 2016, 61(4): 507-519.
pmid: 26876937 |
| [103] |
Li Y, Bedi RK, Wiedmer L, Sun XQ, Huang DZ, Caflisch A. Atomistic and thermodynamic analysis of N6-methyladenosine (m6A) recognition by the reader domain of YTHDC1. J Chem Theory Comput, 2021, 17(2): 1240-1249.
pmid: 33472367 |
| [104] |
Li Y, Bedi RK, Moroz-Omori EV, Caflisch A. Structural and dynamic insights into redundant function of YTHDF proteins. J Chem Inf Model, 2020, 60(12): 5932-5935.
pmid: 33073985 |
| [105] |
Shi HL, Zhang XL, Weng YL, Lu ZY, Liu YJ, Lu ZK, Li JN, Hao PL, Zhang Y, Zhang F, Wu Y, Delgado JY, Su YJ, Patel MJ, Cao XH, Shen B, Huang XX, Ming GL, Zhuang XX, Song HJ, He C, Zhou T. m6A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature, 2018, 563(7730): 249-253.
pmid: 30401835 |
| [106] |
Zhu TT, Roundtree IA, Wang P, Wang X, Wang L, Sun C, Tian Y, Li J, He C, Xu YH. Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine. Cell Res, 2014, 24(12): 1493-1496.
pmid: 25412661 |
| [107] |
Shi HL, Wang X, Lu ZK, Zhao BXS, Ma HH, Hsu PJ, Liu C, He C. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res, 2017, 27(3): 315-328.
pmid: 28106072 |
| [108] |
Zhou WX, Han ZJ, Wu ZX, Gong WK, Yang S, Chen L, Li CH. Specific recognition between YTHDF3 and m6 A-modified RNA: an all-atom molecular dynamics simulation study. Protein, 2022, 90(11): 1965-1972.
pmid: 35639481 |
| [109] |
Hamelberg D, Mongan J, McCammon JA. Accelerated molecular dynamics: a promising and efficient simulation method for biomolecules. J Chem Physs, 2004, 120(24): 11919-11929.
pmid: 15268227 |
| [110] | Li MW, Chen GL, Zhang ZY. Structural and dynamic properties of the YTH domain in complex with N6-methyladenosine RNA studied by accelerated molecular dynamics simulations. Quant Biol, 2023, 11(1): 72-81. |
| [111] |
Piomponi V, Fröhlking T, Bernetti M, Bussi G. Molecular simulations matching denaturation experiments for N6-methyladenosine. ACS Cent Sci, 2022, 8(8): 1218-1228.
pmid: 36032773 |
| [112] |
Piomponi V, Krepl M, Sponer J, Bussi G. Molecular simulations to investigate the impact of N6-methylation in RNA recognition: improving accuracy and precision of binding free energy prediction. J Phys Chem B, 2024, 128(37): 8896-8907.
pmid: 39240243 |
| [113] |
Kierzek E, Zhang XJ, Watson RM, Kennedy SD, Szabat M, Kierzek R, Mathews DH. Secondary structure prediction for RNA sequences including N6-methyladenosine. Nat Commun, 2022, 13(1): 1271.
pmid: 35277476 |
| [114] |
Abramson J, Adler J, Dunger J, Evans R, Green T, Pritzel A, Ronneberger O, Willmore L, Ballard AJ, Bambrick J, Bodenstein SW, Evans DA, Hung CC, O'Neill M, Reiman D, Tunyasuvunakool K, Wu Z, Žemgulytė A, Arvaniti E, Beattie C, Bertolli O, Bridgland A, Cherepanov A, Congreve M, Cowen-Rivers AI, Cowie A, Figurnov M, Fuchs FB, Gladman H, Jain R, Khan YA, Low CMR, Perlin K, Potapenko A, Savy P, Singh S, Stecula A, Thillaisundaram A, Tong C, Yakneen S, Zhong ED, Zielinski M, Žídek A, Bapst V, Kohli P, Jaderberg M, Hassabis D, Jumper JM. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature, 2024, 630(8016): 493-500.
pmid: 38718835 |
| [115] | Penić RJ, Vlašić T, Huber RG, Wan Y, Šikić M. Rinalmo: general-purpose rna language models can generalize well on structure prediction tasks. arXiv, 2024, doi: 10.48550/arXiv.2403.00043. |
| [116] |
Rother M, Rother K, Puton T, Bujnicki JM. ModeRNA: a tool for comparative modeling of RNA 3D structure. Nucleic Acids Res, 2011, 39(10): 4007-4022.
pmid: 21300639 |
| [117] |
Biesiada M, Purzycka KJ, Szachniuk M, Blazewicz J, Adamiak RW. Automated RNA 3D structure prediction with RNAComposer. Methods Mol Biol, 2016, 1490: 199-215.
pmid: 27665601 |
| [118] |
Bernardi RC, Melo MC, Schulten K. Enhanced sampling techniques in molecular dynamics simulations of biological systems. Biochim Biophys Acta, 2015, 1850(5): 872-877.
pmid: 25450171 |
| [119] |
Salekin S, Mostavi M, Chiu YC, Chen Y, Zhang JM, Huang Y. Predicting sites of epitranscriptome modifications using unsupervised representation learning based on generative adversarial networks. Front Phys, 2020, 8: 196.
pmid: 33274189 |
| [120] |
Song ZT, Huang DY, Song BW, Chen KQ, Song YY, Liu G, Su JL, De Magalhães JP, Rigden DJ, Meng J. Attention-based multi-label neural networks for integrated prediction and interpretation of twelve widely occurring RNA modifications. Nat Commun, 2021, 12(1): 4011.
pmid: 34188054 |
| [121] | Wang X, Gu RC, Chen ZY, Li YG, Ji XH, Ke GL, Wen H. UNI-RNA: universal pre-trained models revolutionize RNA research. bioRxiv, 2023, doi: 10.1101/2023.07.11.548588. |
| [122] | Chen JY, Hu ZH, Sun SQ, Tan QX, Wang YX, Yu QZ, Zong LC, Hong L, Xiao J, Shen T, King I, Li Y. Interpretable RNA foundation model from unannotated data for highly accurate RNA structure and function predictions. arXiv, 2022, doi: 10.48550/arXiv.2204.00300. |
| [123] |
Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature, 2009, 462(7271): 315-322.
pmid: 19829295 |
| [124] |
Jeong Y, Gerhauser C, Sauter G, Schlomm T, Rohr K, Lutsik P. MethylBERT enables read-level DNA methylation pattern identification and tumour deconvolution using a Transformer-based model. Nat Commun, 2025, 16(1): 788.
pmid: 39824848 |
| [125] | Li MY, Gu RC, Fan SY, Fan Y, He B, Yang JM, Chen YT, Xin ML, Wen H, Yi CQ. MethylQUEEN: a methylation encoded DNA foundation model. bioRxiv, 2024, doi: 10.1101/2024.12.26.630389. |
| [126] | Marx D, Hutter J. Ab initio molecular dynamics: theory and implementation. Modern Methods and Algorithms of Quantum Chemistry, 2000, 1: 301-449. |
| No related articles found! |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||

www.chinagene.cn
备案号:京ICP备09063187号-4
总访问:,今日访问:,当前在线:
