遗传 ›› 2025, Vol. 47 ›› Issue (8): 885-902.doi: 10.16288/j.yczz.25-052
收稿日期:2025-02-20
修回日期:2025-06-18
出版日期:2025-06-19
发布日期:2025-06-19
通讯作者:
王进,博士,教授,研究方向:RNA生物化学与表观转录组学。E-mail: jinwang@imu.edu.cn;作者简介:冯莹,博士研究生,专业方向:生物化学与分子生物学。E-mail: 1650977020@qq.com冯莹和何晓丽并列第一作者。
基金资助:
Ying Feng( ), Xiaoli He(
), Xiaoli He( ), Yu Liu(
), Yu Liu( ), Jin Wang(
), Jin Wang( )
)
Received:2025-02-20
Revised:2025-06-18
Published:2025-06-19
Online:2025-06-19
Supported by:摘要:
核糖核酸(RNA)是一类关键的生物分子,负责遗传信息的传递、蛋白质的合成及其调控,以及众多生化过程的调节。它们也是许多病毒的关键组成部分。经过化学修饰的合成RNA或寡核糖核苷酸正越来越被广泛地用作治疗药物和疫苗。对于检测、测序、识别和量化RNA及其修饰的技术需求,远远超过了对DNA相关技术的需求。目前,质谱分析法已成为用于识别、测序和量化RNA及其修饰的主要技术方法。本文主要综述了质谱分析法在RNA及其修饰研究中的最新进展,并探讨了该技术方法的优劣势,旨在为读者提供从技术基础到应用前景的全面视角,推动质谱在RNA研究中的更广泛应用,并为领域内方法开发者和生物学研究者提供重要参考。
冯莹, 何晓丽, 刘宇, 王进. 基于质谱的RNA及其修饰分析[J]. 遗传, 2025, 47(8): 885-902.
Ying Feng, Xiaoli He, Yu Liu, Jin Wang. Mass spectrometry-based analysis of RNA and its modifications[J]. Hereditas(Beijing), 2025, 47(8): 885-902.
表1
可用于绘制RNA修饰图谱的酶的切割特异性和局限性"
| 酶 | 切割特异性 | 局限性 | 参考文献 |
|---|---|---|---|
| RNase A | 嘧啶核苷和假尿苷的3′端 | 会生成较短的降解产物,不利于修饰位点的确定 | [ |
| hRNase 4 | 位于嘌呤5′端且与之相邻的尿苷 | 适用于mRNA和其他长链RNA,但需要添加T4多核苷酸激酶以避免生成2′,3′-环磷酸酯 | [ |
| RNase T1 | 鸟苷和N2-甲基鸟苷(m2G)的3′端 | 无法生成高的序列覆盖度,尤其是在存在富含G的序列冗余时 | [ |
| MC1 | 尿苷和假尿苷的5′端 | 不能区分假尿苷和尿苷 | [ |
| Cusativin | 胞苷和m5C的5′端 | 连续胞苷残基之间没有磷酸二酯键切割 | [ |
| RNase H | DNA-RNA杂合链中的RNA | 可能产生非特异性切割 | [ |
| RNase U2 | 嘌呤残基处切割RNA | 生成2′,3′-环磷酸酯和3′-线性磷酸酯消化产物,使得质谱分析变得复杂 | [ |
| colicin E5 | 在某些tRNA的第34位的辫苷(Q)与第35位的尿苷之间切割 | 仅对大肠杆菌的特定tRNA具有切割作用 | [ |
| mazF | 在5′-NAC-3′序列(N优先为U或A)中的N与A之间切割 | 对RNA底物的识别和切割活性易受RNA二级结构的影响 | [ |
表2
可用于绘制RNA修饰图谱的软件程序"
| 软件 | 优点 | 缺点 | 参考文献 |
|---|---|---|---|
| NIST spectral software | 可鉴定特定修饰的存在或不存在 | 不能区分尿苷和胞苷; 只能用于已知的寡核苷酸 | [ |
| RAMM | 计算机(in silico)数据库; 固定和可变测序 | 不能区分相同经典核苷的甲基化修饰和硫修饰; 不能完全消除对数据的人工解析 | [ |
| RoboOligo | 复杂修饰和多个修饰的从头测序 | 不能区分核苷位置异构体; 不能区分m/z值相同或几乎相同的母离子; 不能区分尿苷和假尿苷 | [ |
| NASE | 开源软件; 校正母离子质量及阳离子加合; 基于目标/诱饵(target/decoy)搜索策略的错误发现率(FDR)参数 | 高错误发现率; 不能完全消除对数据的人工解析; 不能区分核苷位置异构体; 不能区分尿苷和假尿苷 | [ |
| Pytheas | 开源软件; 适用于同位素标记序列; 基于目标/诱饵搜索策略的FDR 能够区分位置异构体; 能够区分尿苷和假尿苷 | 需要人工解析数据; 需要大量计算 | [ |
| Ariadne | RNA鉴定和测序自动化 | 缺乏通过FDR计算对匹配输出结果进行验证; 不能区分核苷位置异构体; 不能区分尿苷和假尿苷 | [ |
| Nucleo-SAFARI | 计算机(in silico)数据库; 自动注释在轨道阱质谱仪平台上记录的核酸top-down MS/MS谱图; 基于R语言的核酸MS/MS谱图分析; 直接基于m/z识别碎片,而无需依赖去卷积和去同位素算法; 能够区分位置异构体 | 不能对多个质谱图和序列进行批量处理 | [ |
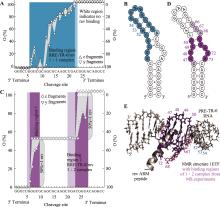
图4
通过CAD MS绘制RRE II RNA构建体RRE-TR-0/ HIV rev复合物的结合位点图谱[137] A:1:1复合物离子 (RRE-TR-0 + 1·rev − 14H)14−在137.2 eV的CAD中c片段(左侧轴)和y片段(右侧轴)的rev ARM肽位点特异性占据率(O),以及映射到预测的RRE-TR-0二级结构B:上的相应结合区域(蓝色),与NMR结构中的结合位点E的一致性较差。C:1:2复合物离子(RRE-TR-0 + 2·rev − 14H)14−在175.5 eV的CAD中的片段的占据率,以及映射到RRE-TR-0结构D上的相应结合位点(紫色),与NMR结构E中的结合位点的一致性较好。B、D和E中的深色和浅色分别代表较强和较弱的结合,与A和C中的深色和浅色背景色相对应。B、D和E中的数字表示序列位点。CAD,碰撞激活解离;MS,质谱;ARM,富含精氨酸基序;NMR,核磁共振。"
| [1] |
Höfer K, Jäschke A. Epitranscriptomics: RNA modifications in bacteria and archaea. Microbiol Spectr, 2018, 6(3): 10.1128/microbiolspec.rwr-0015-2017.
pmid: 29916347 |
| [2] |
Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell, 2017, 169(7): 1187-1200.
pmid: 28622506 |
| [3] |
Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, de Crécy-Lagard V, Ross R, Limbach PA, Kotter A, Helm M, Bujnicki JM. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Research, 2018, 46(D1): D303-D307.
pmid: 29106616 |
| [4] |
Cappannini A, Ray A, Purta E, Mukherjee S, Boccaletto P, Moafinejad SN, Lechner A, Barchet C, Klaholz BP, Stefaniak F, Bujnicki JM. MODOMICS: a database of RNA modifications and related information. 2023 update. Nucleic Acids Res, 2024, 52(D1): D239-D244.
pmid: 38015436 |
| [5] |
Wang J, Dong HP, Chionh YH, McBee ME, Sirirungruang S, Cunningham RP, Shi PY, Dedon PC. The role of sequence context, nucleotide pool balance and stress in 2′-deoxynucleotide misincorporation in viral, bacterial and mammalian RNA. Nucleic Acids Res, 2016, 44(18): 8962-8975.
pmid: 27365049 |
| [6] |
Sekiguchi T, Ito R, Hayakawa H, Sekiguchi M. Elimination and utilization of oxidized guanine nucleotides in the synthesis of RNA and its precursors. J Biol Chem, 2013, 288(12): 8128-8135.
pmid: 23376345 |
| [7] | Hong CD, Chi Z, Jia GF. The biological regulation of RNA modifications. Chin Bulletin of Life Sciences, 2018, 30(04): 414-423. |
| 段洪超, 张弛, 贾桂芳. RNA修饰的生物学功能. 生命科学, 2018, 30(4): 414-423. | |
| [8] |
Kong QM, Lin CLG. Oxidative damage to RNA: mechanisms, consequences, and diseases. Cell Mol Life Sci, 2010, 67(11): 1817-1829.
pmid: 20148281 |
| [9] |
Boulias K, Greer EL. Biological roles of adenine methylation in RNA. Nat Rev Genet, 2023, 24(3): 143-160.
pmid: 36261710 |
| [10] |
Lin SB, Kuang M. RNA modification-mediated mRNA translation regulation in liver cancer: mechanisms and clinical perspectives. Nat Rev Gastroenterol Hepatol, 2024, 21(4): 267-281.
pmid: 38243019 |
| [11] |
Simms CL, Zaher HS. Quality control of chemically damaged RNA. Cell Mol Life Sci, 2016, 73(19): 3639-3653.
pmid: 27155660 |
| [12] |
Delaunay S, Pascual G, Feng B, Klann K, Behm M, Hotz-Wagenblatt A, Richter K, Zaoui K, Herpel E, Münch C, Dietmann S, Hess J, Benitah SA, Frye M. Mitochondrial RNA modifications shape metabolic plasticity in metastasis. Nature, 2022, 607(7919): 593-603.
pmid: 35768510 |
| [13] |
Li R, Zhao HZ, Huang XD, Zhang JL, Bai RH, Zhuang LS, Wen SJ, Wu SJ, Zhou QB, Li M, Zeng LX, Zhang SP, Deng S, Su JC, Zuo ZX, Chen RF, Lin DX, Zheng J. Super-enhancer RNA m6A promotes local chromatin accessibility and oncogene transcription in pancreatic ductal adenocarcinoma. Nat Genet, 2023, 55(12): 2224-2234.
pmid: 37957340 |
| [14] |
Delaunay S, Helm M, Frye M. RNA modifications in physiology and disease: towards clinical applications. Nat Rev Genet, 2024, 25(2): 104-122.
pmid: 37714958 |
| [15] |
Yang L, Ma ML, Gao YW, Liu J. Decoding N6-methyladenosine's dynamic role in stem cell fate and early embryo development: insights into RNA-chromatin interactions. Curr Opin Genet Dev, 2025, 91: 102311.
pmid: 39908649 |
| [16] |
Tang SL, Liu GY, Yan YT, Wang SH, Li N. Development of a flow through-based limited digestion approach for high-throughput and high-sequence coverage mapping of therapeutic mRNAs. Anal Chem, 2024, 96(42): 16994-17003.
pmid: 39391985 |
| [17] |
Wang J, Chew BLA, Lai Y, Dong HP, Xu L, Liu Y, Fu XY, Lin ZG, Shi PY, Lu TK, Luo DH, Jaffrey SR, Dedon PC. A systems-level mass spectrometry-based technique for accurate and sensitive quantification of the RNA cap epitranscriptome. Nat Protoc, 2023, 18(9): 2671-2698.
pmid: 37567932 |
| [18] |
Jain M, Abu-Shumays R, Olsen HE, Akeson M. Advances in nanopore direct RNA sequencing. Nat Methods, 2022, 19(10): 1160-1164.
pmid: 36203024 |
| [19] |
Su D, Chan CTY, Gu C, Lim KS, Chionh YH, McBee ME, Russell BS, Babu IR, Begley TJ, Dedon PC. Quantitative analysis of ribonucleoside modifications in tRNA by HPLC-coupled mass spectrometry. Nat Protoc, 2014, 9(4): 828-841.
pmid: 24625781 |
| [20] | Xiao PC, Run Z, Yuan-mei W, Huan-ming Y. The RNome Project: Another Human Genome Project. Hereditas (Beijing), 2024, 46(5): 357-359. |
| 岑萧萍, 周润, 王元梅, 杨焕明. RNome计划: 又一个人类基因组计划. 遗传, 2024, 46(5): 357-359. | |
| [21] |
Stephenson W, Razaghi R, Busan S, Weeks KM, Timp W, Smibert P. Direct detection of RNA modifications and structure using single-molecule nanopore sequencing. Cell Genom, 2022, 2(2): 100097.
pmid: 35252946 |
| [22] |
Akaçin İ, Ersoy Ş, Doluca O, Güngörmüşler M. Comparing the significance of the utilization of next generation and third generation sequencing technologies in microbial metagenomics. Microbiol Res, 2022, 264: 127154.
pmid: 35961096 |
| [23] |
Deng L, Kumar J, Rose R, McIntyre W, Fabris D. Analyzing RNA posttranscriptional modifications to decipher the epitranscriptomic code. Mass Spectrom Rev, 2024, 43(1): 5-38.
pmid: 36052666 |
| [24] |
Sarkar A, Gasperi W, Begley U, Nevins S, Huber SM, Dedon PC, Begley TJ. Detecting the epitranscriptome. Wiley Interdiscip Rev RNA, 2021, 12(6): e1663.
pmid: 33987958 |
| [25] |
Heiss M, Borland K, Yoluç Y, Kellner S. Quantification of modified nucleosides in the context of nail-ms. Methods Mol Biol, 2021, 2298: 279-306.
pmid: 34085252 |
| [26] |
Thüring K, Schmid K, Keller P, Helm M. Analysis of RNA modifications by liquid chromatography-tandem mass spectrometry. Methods, 2016, 107: 48-56.
pmid: 27020891 |
| [27] | Lan MD, Yuan BF, Feng YQ. Deciphering nucleic acid modifications by chemical derivatization-mass spectrometry analysis. Chin Chem Lett, 2019, 30(1): 1-6. |
| [28] |
Xiong J, Yuan BF, Feng YQ. Mass spectrometry for investigating the effects of toxic metals on nucleic acid modifications. Chem Res Toxicol, 2019, 32(5): 808-819.
pmid: 30920205 |
| [29] | Tao WB, Xie NB, Cheng QY, Feng YQ, Yuan BF. Sensitive determination of inosine RNA modification in single cell by chemical derivatization coupled with mass spectrometry analysis. Chin Chem Lett, 2023, 34(10): 108243. |
| [30] |
Zhang HY, Xiong J, Qi BL, Feng YQ, Yuan BF. The existence of 5-hydroxymethylcytosine and 5-formylcytosine in both DNA and RNA in mammals. Chem Commun (Camb), 2016, 52(4): 737-740.
pmid: 26562407 |
| [31] | Xiong J, Wu JY, Liu Y, Feng YJ, Yuan BF. Quantification and mapping of RNA modifications. TrAC Trends Anal Chem, 2024, 172: 117606. |
| [32] |
Dai Y, Qi CB, Feng Y, Cheng QY, Liu FL, Cheng MY, Yuan BF, Feng YQ. Sensitive and simultaneous determination of uridine thiolation and hydroxylation modifications in eukaryotic RNA by derivatization coupled with mass spectrometry analysis. Anal Chem, 2021, 93(18): 6938-6946.
pmid: 33908769 |
| [33] |
Huang W, Lan MD, Qi CB, Zheng SJ, Wei SZ, Yuan BF, Feng YQ. Formation and determination of the oxidation products of 5-methylcytosine in RNA. Chem Sci, 2016, 7(8): 5495-5502.
pmid: 30034689 |
| [34] |
Xie YX, Janssen KA, Scacchetti A, Porter EG, Lin ZT, Bonasio R, Garcia BA. Permethylation of ribonucleosides provides enhanced mass spectrometry quantification of post-transcriptional RNA modifications. Anal Chem, 2022, 94(20): 7246-7254.
pmid: 35549217 |
| [35] |
Quinn R, Basanta-Sanchez M, Rose RE, Fabris D. Direct infusion analysis of nucleotide mixtures of very similar or identical elemental composition. J Mass Spectrom, 2013, 48(6): 703-712.
pmid: 23722961 |
| [36] |
Rose RE, Quinn R, Sayre JL, Fabris D. Profiling ribonucleotide modifications at full-transcriptome level: a step toward MS-based epitranscriptomics. RNA, 2015, 21(7): 1361-1374.
pmid: 25995446 |
| [37] |
Jones J D, Simcox K M, Kennedy R T, Koutmou KS. Direct sequencing of total saccharomyces cerevisiae tRNAs by LC-MS/MS. RNA, 2023, 29(8): 1201-1214.
pmid: 37169396 |
| [38] | Popova A M, Williamson J R. Quantitative analysis of rRNA modifications using stable isotope labeling and mass spectrometry. J Am Chem Soc, 2014, 136(5): 2058-2069. |
| [39] |
Tardu M, Jones J D, Kennedy R T, Lin Q, Koutmou K S. Identification and quantification of modified nucleosides in saccharomyces cerevisiae mRNAs. ACS Chem Biol, 2019, 14(7): 1403-1409.
pmid: 31243956 |
| [40] |
Feng YJ, You XJ, Ding JH, Zhang YF, Yuan BF, Feng YQ. Identification of inosine and 2′-O-methylinosine modifications in yeast messenger RNA by liquid chromatography-tandem mass spectrometry analysis. Anal Chem, 2022, 94(11): 4747-4755.
pmid: 35266699 |
| [41] |
Petrov DP, Kaiser S, Kaiser S, Jung K. Opportunities and challenges to profile mRNA modifications in escherichia coli. Chembiochem, 2022, 23(18): e202200270.
pmid: 35822398 |
| [42] |
Chan CTY, Dyavaiah M, DeMott MS, Taghizadeh K, Dedon PC, Begley TJ. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet, 2010, 6(12): e1001247.
pmid: 21187895 |
| [43] |
Dedon PC, Begley TJ. Dysfunctional tRNA reprogramming and codon-biased translation in cancer. Trends Mol Med, 2022, 28(11): 964-978.
pmid: 36241532 |
| [44] |
Huber SM, Leonardi A, Dedon PC, Begley TJ. The versatile roles of the tRNA epitranscriptome during cellular responses to toxic exposures and environmental stress. Toxics, 2019, 7(1): 17.
pmid: 30934574 |
| [45] |
Endres L, Dedon PC, Begley TJ. Codon-biased translation can be regulated by wobble-base tRNA modification systems during cellular stress responses. RNA Biol, 2015, 12(6): 603-614.
pmid: 25892531 |
| [46] |
Gu C, Begley TJ, Dedon PC. tRNA modifications regulate translation during cellular stress. FEBS Lett, 2014, 588(23): 4287-4296.
pmid: 25304425 |
| [47] |
Yoluç Y, van de Logt E, Kellner-Kaiser S. The stress-dependent dynamics of saccharomyces cerevisiae tRNA and rRNA Modification Profiles. Genes (Basel), 2021, 12(9): 1344.
pmid: 34573326 |
| [48] |
Heiss M, Hagelskamp F, Marchand V, Motorin Y, Kellner S. Cell culture NAIL-MS allows insight into human tRNA and rRNA modification dynamics in vivo. Nat Commun, 2021, 12(1): 389.
pmid: 33452242 |
| [49] |
Herbert C, Valesyan S, Kist J, Limbach PA. Analysis of RNA and its modifications. Annu Rev Anal Chem (Palo Alto Calif), 2024, 17(1): 47-68.
pmid: 38594935 |
| [50] |
Wang J, Alvin Chew B L, Lai Y, Dong HP, Xu L, Balamkundu S, Cai WM, Cui L, Liu CF, Fu XY, Lin ZG, Shi PY, Lu TK, Luo DH, Jaffrey SR, Dedon PC. Quantifying the RNA cap epitranscriptome reveals novel caps in cellular and viral RNA. Nucleic Acids Res, 2019, 47(20): e130.
pmid: 31504804 |
| [51] |
Espadas G, Morales-Sanfrutos J, Medina R, Lucas MC, Novoa EM, Sabidó E. High-performance nano-flow liquid chromatography column combined with high- and low-collision energy data-independent acquisition enables targeted and discovery identification of modified ribonucleotides by mass spectrometry. J Chromatogr A, 2022, 1665: 462803.
pmid: 35042139 |
| [52] |
Jora M, Burns AP, Ross RL, Lobue PA, Zhao RX, Palumbo CM, Beal PA, Addepalli B, Limbach PA. Differentiating positional Isomers of nucleoside modifications by higher-energy collisional dissociation mass spectrometry (hcd ms). J Am Soc Mass Spectrom, 2018, 29(8): 1745-1756.
pmid: 29949056 |
| [53] |
Li YR, Zhou J, Yuan G. Discrimination of common isomerides of methyl nucleosides by collision-induced dissociation tandem mass spectrometry. J Mass Spectrom, 2020, 56(4): e4594.
pmid: 32639684 |
| [54] |
Macon JB, Wolfenden R. 1-Methyladenosine. Dimroth rearrangement and reversible reduction. Biochemistry, 1968, 7(10): 3453-3458.
pmid: 5681457 |
| [55] |
Miyauchi K, Kimura S, Suzuki T. A cyclic form of N6-threonylcarbamoyladenosine as a widely distributed tRNA hypermodification. Nat Chem Biol, 2013, 9(2): 105-111.
pmid: 23242255 |
| [56] |
Ammann G, Berg M, Dalwigk JF, Kaiser SM. Pitfalls in RNA modification quantification using nucleoside mass Spectrometry. Acc Chem Res, 2023, 56(22): 3121-3131.
pmid: 37944919 |
| [57] |
Mair S, Erharter K, Renard E, Brillet K, Brunner M, Lusser A, Kreutz C, Ennifar E, Micura R. Towards a comprehensive understanding of RNA deamination: synthesis and properties of xanthosine-modified RNA. Nucleic Acids Res, 2022, 50(11): 6038-6051.
pmid: 35687141 |
| [58] |
Jora M, Borland K, Abernathy S, Zhao RX, Kelley M, Kellner S, Addepalli B, Limbach PA. Chemical amination/imination of carbonothiolated nucleosides during RNA hydrolysis. Angew Chem Int Ed Engl, 2021, 60(8): 3961-3966.
pmid: 33125801 |
| [59] |
Havelund JF, Giessing AMB, Hansen T, Rasmussen A, Scott LG, Kirpekar F. Identification of 5-hydroxycytidine at position 2501 concludes characterization of modified nucleotides in E. coli 23S rRNA. J Mol Biol, 2011, 411(3): 529-536.
pmid: 21723290 |
| [60] |
Taghizadeh K, McFaline JL, Pang B, Sullivan M, Dong M, Plummer E, Dedon PC. Quantification of DNA damage products resulting from deamination, oxidation and reaction with products of lipid peroxidation by liquid chromatography isotope dilution tandem mass spectrometry. Nat Protoc, 2008, 3(8): 1287-1298.
pmid: 18714297 |
| [61] |
Richter F, Plehn JE, Bessler L, Hertler J, Jörg M, Cirzi C, Tuorto F, Friedland K, Helm M. RNA marker modifications reveal the necessity for rigorous preparation protocols to avoid artifacts in epitranscriptomic analysis. Nucleic Acids Res, 2022, 50(8): 4201-4215.
pmid: 34850949 |
| [62] |
Jora M, Corcoran D, Parungao GG, Lobue PA, Oliveira LFL, Stan G, Addepalli B, Limbach PA. Higher- energy collisional dissociation mass spectral networks for the rapid, semi-automated characterization of known and unknown ribonucleoside modifications. Anal Chem, 2022, 94(40): 13958-13967.
pmid: 36174068 |
| [63] |
Gosset-Erard C, Didierjean M, Pansanel J, Lechner A, Wolff P, Kuhn L, Aubriet F, Leize-Wagner E, Chaimbault P, François YN. Nucleos'ID: a new search engine enabling the untargeted identification of RNA post- transcriptional modifications from tandem mass spectrometry analyses of nucleosides. Anal Chem, 2023, 95(2): 1608-1617.
pmid: 36598775 |
| [64] |
Roberts TC, Langer R, Wood MJA. Advances in oligonucleotide drug delivery. Nat Rev Drug Discov, 2020, 19(10): 673-694.
pmid: 32782413 |
| [65] |
Khvorova A, Watts JK. The chemical evolution of oligonucleotide therapies of clinical utility. Nat Biotechnol, 2017, 35(3): 238-248.
pmid: 28244990 |
| [66] |
Apffel A, Chakel JA, Fischer S, Lichtenwalter K, Hancock WS. Analysis of oligonucleotides by HPLC- electrospray Ionization Mass Spectrometry. Anal Chem, 1997, 69(7): 1320-1325.
pmid: 21639339 |
| [67] |
Studzińska S, Bocian S, Siecińska L, Buszewski B. Application of phenyl-based stationary phases for the study of retention and separation of oligonucleotides. J Chromatogr B Analyt Technol Biomed Life Sci, 2017, 1060: 36-43.
pmid: 28595118 |
| [68] |
Bartlett MG, Omuro S. Evaluation of alkylamines and stationary phases to improve LC-MS of oligonucleotides. Biomed Chromatogr, 2021, 35(5): e5045.
pmid: 33283300 |
| [69] |
McGinnis AC, Grubb EC, Bartlett MG. Systematic optimization of ion-pairing agents and hexafluoroisopropanol for enhanced electrospray ionization mass spectrometry of oligonucleotides. Rapid Commun Mass Spectrom, 2013, 27(23): 2655-2664.
pmid: 24591027 |
| [70] |
Chen XC, Liu ZQ, Gong LZ. Evaluating the interplay among stationary phases/ion-pairing reagents/sequences for liquid chromatography mass spectrometry analysis of oligonucleotides. Anal Biochem, 2021, 625: 114194.
pmid: 33910045 |
| [71] |
Studzińska S, Cywoniuk P, Sobczak K. Application of ion pair chromatography coupled with mass spectrometry to assess antisense oligonucleotides concentrations in living cells. Analyst, 2019, 144(2): 622-633.
pmid: 30462105 |
| [72] |
Li N, El Zahar NM, Saad JG, van der Hage ERE, Bartlett MG. Alkylamine ion-pairing reagents and the chromatographic separation of oligonucleotides. J Chromatogr A, 2018, 1580: 110-119.
pmid: 30409418 |
| [73] |
Donegan M, Nguyen JM, Gilar M. Effect of ion-pairing reagent hydrophobicity on liquid chromatography and mass spectrometry analysis of oligonucleotides. J Chromatogr A, 2022, 1666: 462860.
pmid: 35123169 |
| [74] |
Enmark M, Harun S, Samuelsson J, Örnskov E, Thunberg L, Dahlén A, Fornstedt T. Selectivity limits of and opportunities for ion pair chromatographic separation of oligonucleotides. J Chromatogr A, 2021, 1651: 462269.
pmid: 34102400 |
| [75] |
Enmark M, Samuelsson J, Fornstedt T. Development of a unified gradient theory for ion-pair chromatography using oligonucleotide separations as a model case. J Chromatogr A, 2023, 1691: 463823.
pmid: 36716595 |
| [76] |
Lobue PA, Jora M, Addepalli B, Limbach PA. Oligonucleotide analysis by hydrophilic interaction liquid chromatography-mass spectrometry in the absence of ion-pair reagents. J Chromatogr A, 2019, 1595: 39-48.
pmid: 30772056 |
| [77] |
Hagelskamp F, Borland K, Ramos J, Hendrick AG, Fu D, Kellner S. Broadly applicable oligonucleotide mass spectrometry for the analysis of RNA writers and erasers in vitro. Nucleic Acids Res, 2020, 48(7): e41.
pmid: 32083657 |
| [78] |
Yuan L, Dupuis JF, Mekhssian K. A novel hybridization LC-MS/MS methodology for quantification of siRNA in plasma, CSF and tissue samples. Molecules, 2023, 28(4): 1618.
pmid: 36838605 |
| [79] |
Guimaraes G, Yuan L, Li P. Antisense oligonucleotide in vitro protein binding determination in plasma, brain, and cerebral spinal fluid using hybridization LC-MS/MS. Drug Metab Dispos, 2022, 50(3): 268-276.
pmid: 34921096 |
| [80] |
Mahajan S, Zhao H, Kovacina K, Lachacz E, Hoxha S, Chan J, Liang M. High-sensitivity quantification of antisense oligonucleotides for pharmacokinetic characterization. Bioanalysis, 2022, 14(9): 603-613.
pmid: 35578971 |
| [81] |
Li P, Gong YQ, Kim J, Liu XR, Gilbert J, Kerns HM, Groth R, Rooney M. Hybridization liquid chromatography- tandem mass spectrometry: an alternative bioanalytical method for antisense oligonucleotide quantitation in plasma and tissue samples. Anal Chem, 2020, 92(15): 10548-10559.
pmid: 32628461 |
| [82] |
Demelenne A, Servais AC, Crommen J, Fillet M. Analytical techniques currently used in the pharmaceutical industry for the quality control of RNA-based therapeutics and ongoing developments. J Chromatogr A, 2021, 1651: 462283.
pmid: 34107400 |
| [83] |
Guimaraes GJ, Sutton JM, Gilar M, Donegan M, Bartlett MG. Impact of nonspecific adsorption to metal surfaces in ion pair-RPLC-MS impurity analysis of oligonucleotides. J Pharm Biomed Anal, 2022, 208: 114439.
pmid: 34742118 |
| [84] |
Roussis SG, Cedillo I, Rentel C. Semi-quantitative determination of co-eluting impurities in oligonucleotide drugs using ion-pair reversed-phase liquid chromatography mass spectrometry. J Chromatogr A, 2019, 1584: 106-114.
pmid: 30473112 |
| [85] |
Pourshahian S. Therapeutic oligonucleotides, impurities, degradants, and their characterization by mass spectrometry. Mass Spectrom Rev, 2021, 40(2): 75-109.
pmid: 31840864 |
| [86] |
Jiang D, Yuan L. Microflow LC-MS/MS to improve sensitivity for antisense oligonucleotides bioanalysis: critical role of sample cleanness. Bioanalysis, 2022, 14(21): 1365-1376.
pmid: 36625771 |
| [87] |
Solivio B, Yu NX, Addepalli B, Limbach PA. Improving RNA modification mapping sequence coverage by LC-MS through a nonspecific RNase U2-E49A mutant. Anal Chim Acta, 2018, 1036: 73-79.
pmid: 30253839 |
| [88] |
Douthwaite S, Kirpekar F. Identifying modifications in RNA by MALDI mass spectrometry. Methods Enzymol, 2007, 425: 3-20.
pmid: 17673077 |
| [89] |
Herbert C, Dzowo YK, Urban A, Kiggins CN, Resendiz MJE. Reactivity and dpecificity of RNase T1, RNase A, and RNase H toward oligonucleotides of RNA containing 8-Oxo-7,8-dihydroguanosine. Biochemistry, 2018, 57(20): 2971-2983.
pmid: 29683663 |
| [90] |
Durairaj A, Limbach PA. Improving CMC-derivatization of pseudouridine in RNA for mass spectrometric detection. Anal Chim Acta, 2008, 612(2): 173-181.
pmid: 18358863 |
| [91] |
Addepalli B, Lesner NP, Limbach PA. Detection of RNA nucleoside modifications with the uridine-specific ribonuclease MC1 from momordica charantia. RNA, 2015, 21(10): 1746-1756.
pmid: 26221047 |
| [92] |
Thakur P, Estevez M, Lobue PA, Limbach PA, Addepalli B. Improved RNA modification mapping of cellular non-coding RNAs using C- and U-specific RNases. Analyst, 2020, 145(3): 816-827.
pmid: 31825413 |
| [93] |
Jiang T, Yu NX, Kim J, Murgo JR, Kissai M, Ravichandran K, Miracco EJ, Presnyak V, Hua S. Oligonucleotide sequence mapping of Large therapeutic mRNAs via parallel ribonuclease digestions and LC-MS/MS. Anal Chem, 2019, 91(13): 8500-8506.
pmid: 31129964 |
| [94] |
Vanhinsbergh CJ, Criscuolo A, Sutton JN, Murphy K, Williamson AJK, Cook K, Dickman MJ. Characterization and sequence mapping of large RNA and mRNA therapeutics using mass spectrometry. Anal Chem, 2022, 94(20): 7339-7349.
pmid: 35549087 |
| [95] |
Pomerantz SC, Kowalak JA, McCloskey JA. Determination of oligonucleotide composition from mass spectrometrically measured molecular weight. J Am Soc Mass Spectrom, 1993, 4(3): 204-209.
pmid: 24234848 |
| [96] |
de Crécy-Lagard V, Ross RL, Jaroch M, Marchand V, Eisenhart C, Brégeon D, Motorin Y, Limbach PA. Survey and validation of tRNA modifications and their corresponding genes in bacillus subtilis sp subtilis strain 168. Biomolecules, 2020, 10(7): 977.
pmid: 32629984 |
| [97] |
Kimura S, Dedon PC, Waldor MK. Comparative tRNA sequencing and RNA mass spectrometry for surveying tRNA modifications. Nat Chem Biol, 2020, 16(9): 964-972.
pmid: 32514182 |
| [98] |
Yan TM, Pan Y, Yu ML, Hu K, Cao KY, Jiang ZH. Full-range profiling of tRNA modifications using LC-MS/MS at single-base resolution through a site- specific cleavage strategy. Anal Chem, 2021, 93(3): 1423-1432.
pmid: 33382261 |
| [99] |
Puri P, Wetzel C, Saffert P, Gaston KW, Russell SP, Cordero Varela JA, van der Vlies P, Zhang G, Limbach PA, Ignatova Z, Poolman B. Systematic identification of tRNAome and its dynamics in Lactococcus lactis. Mol Microbiol, 2014, 93(5): 944-956.
pmid: 25040919 |
| [100] |
Prats-Ejarque G, Lu L, Salazar VA, Moussaoui M, Boix E. Evolutionary trends in RNA base selectivity within the RNase A superfamily. Front Pharmacol, 2019, 10: 1170.
pmid: 31649540 |
| [101] |
Wolf EJ, Grünberg S, Dai N, Chen TH, Roy B, Yigit E, Corrêa IR. Human RNase 4 improves mRNA sequence characterization by LC-MS/MS. Nucleic Acids Res, 2022, 50(18): e106.
pmid: 35871301 |
| [102] |
Greiner-Stöffele T, Foerster HH, Hahn U. Ribonuclease T1 cleaves RNA after guanosines within single-stranded gaps of any length. Nucleosides Nucleotides Nucleic Acids, 2000, 19(7): 1101-1109.
pmid: 10999250 |
| [103] |
Addepalli B, Venus S, Thakur P, Limbach PA. Novel ribonuclease activity of cusativin from Cucumis sativus for mapping nucleoside modifications in RNA. Anal Bioanal Chem, 2017, 409(24): 5645-5654.
pmid: 28730304 |
| [104] |
Beverly M, Dell A, Parmar P, Houghton L. Label-free analysis of mRNA capping efficiency using RNase H probes and LC-MS. Anal Bioanal Chem, 2016, 408(18): 5021-5030.
pmid: 27193635 |
| [105] |
Inoue H, Hayase Y, Iwai S, Ohtsuka E. Sequence- dependent hydrolysis of RNA using modified oligonucleotide splints and RNase H. FEBS Lett, 1987, 215(2): 327-330.
pmid: 2438160 |
| [106] |
Chan SH, Whipple JM, Dai N, Kelley TM, Withers K, Tzertzinis G, Corrêa IR, Jr., Robb GB. RNase H-based analysis of synthetic mRNA 5′ cap incorporation. RNA, 2022, 28(8): 1144-1155.
pmid: 35680168 |
| [107] |
Houser WM, Butterer A, Addepalli B, Limbach PA. Combining recombinant ribonuclease U2 and protein phosphatase for RNA modification mapping by liquid chromatography-mass spectrometry. Anal Biochem, 2015, 478: 52-58.
pmid: 25797349 |
| [108] |
Ogawa T, Inoue S, Yajima S, Hidaka M, Masaki H. Sequence-specific recognition of colicin E5, a tRNA-targeting ribonuclease. Nucleic Acids Res, 2006, 34(21): 6065-6073.
pmid: 16963495 |
| [109] |
Yajima S, Inoue S, Ogawa T, Nonaka T, Ohsawa K, Masaki H. Structural basis for sequence-dependent recognition of colicin E5 tRNase by mimicking the mRNA-tRNA interaction. Nucleic Acids Res, 2006, 34(21): 6074-6082.
pmid: 17099236 |
| [110] |
Muñoz-Gómez AJ, Santos-Sierra S, Berzal-Herranz A, Lemonnier M, Díaz-Orejas R. Insights into the specificity of RNA cleavage by the Escherichia coli MazF toxin. FEBS Lett, 2004, 567(2-3): 316-320.
pmid: 15178344 |
| [111] |
Catherman AD, Skinner OS, Kelleher NL. Top Down proteomics: facts and perspectives. Biochem Biophys Res Commun, 2014, 445(4): 683-693.
pmid: 24556311 |
| [112] |
Siuti N, Kelleher NL. Decoding protein modifications using top-down mass spectrometry. Nat Methods, 2007, 4(10): 817-821.
pmid: 17901871 |
| [113] |
Jebanathirajah JA, Pittman JL, Thomson BA, Budnik BA, Kaur P, Rape M, Kirschner M, Costello CE, O'Connor PB. Characterization of a new qQq-FTICR mass spectrometer for post-translational modification analysis and top-down tandem mass spectrometry of whole proteins. J Am Soc Mass Spectrom, 2005, 16(12): 1985-1999.
pmid: 16271296 |
| [114] |
Huang TY, Liu J, McLuckey SA. Top-down tandem mass spectrometry of tRNA via ion trap collision-induced dissociation. J Am Soc Mass Spectrom, 2010, 21(6): 890-898.
pmid: 20080046 |
| [115] |
Riml C, Glasner H, Rodgers MT, Micura R, Breuker K. On the mechanism of RNA phosphodiester backbone cleavage in the absence of solvent. Nucleic Acids Res, 2015, 43(10): 5171-5181.
pmid: 25904631 |
| [116] |
Taucher M, Breuker K. Top-down mass spectrometry for sequencing of larger (up to 61 nt) RNA by CAD and EDD. J Am Soc Mass Spectrom, 2010, 21(6): 918-929.
pmid: 20363646 |
| [117] |
Taucher M, Breuker K. Characterization of modified RNA by top-down mass spectrometry. Angew Chem Int Ed Engl, 2012, 51(45): 11289-11292.
pmid: 23042528 |
| [118] |
Glasner H, Riml C, Micura R, Breuker K. Label-free. direct localization and relative quantitation of the RNA nucleobase methylations m6A, m5C, m3U, and m5U by top-down mass spectrometry. Nucleic Acids Res, 2017, 45(13): 8014-8025.
pmid: 28549193 |
| [119] |
Peters-Clarke TM, Quan QW, Brademan DR, Hebert AS, Westphall MS, Coon JJ. Ribonucleic acid sequence characterization by negative electron transfer dissociation mass spectrometry. Anal Chem, 2020, 92(6): 4436-4444.
pmid: 32091202 |
| [120] |
Santos IC, Lanzillotti M, Shilov I, Basanta-Sanchez M, Roushan A, Lawler R, Tang W, Bern M, Brodbelt JS. Ultraviolet photodissociation and activated electron photodetachment mass spectrometry for top-down sequencing of modified oligoribonucleotides. J Am Soc Mass Spectrom, 2022, 33(3): 510-520.
pmid: 35157441 |
| [121] |
Mutchek S, Kenderdine T, Turner K, Ring J, German M, Fabris D. Strand-cleaving deoxyribozymes enable the mid-down sequencing of mRNA by mass spectrometry with no front-end separations. Anal Chem, 2025, 97(5): 2972-2980.
pmid: 39888840 |
| [122] |
Gaston KW, Limbach PA. The identification and characterization of non-coding and coding RNAs and their modified nucleosides by mass spectrometry. RNA Biol, 2014, 11(12): 1568-1585.
pmid: 25616408 |
| [123] |
D'Ascenzo L, Popova AM, Abernathy S, Sheng K, Limbach PA, Williamson JR. Pytheas: a software package for the automated analysis of RNA sequences and modifications via tandem mass spectrometry. Nat Commun, 2022, 13(1): 2424.
pmid: 35505047 |
| [124] |
Wein S, Andrews B, Sachsenberg T, Santos-Rosa H, Kohlbacher O, Kouzarides T, Garcia BA, Weisser H. A computational platform for high-throughput analysis of RNA sequences and modifications by mass spectrometry. Nat Commun, 2020, 11(1): 926.
pmid: 32066737 |
| [125] |
Yu NX, Lobue PA, Cao XY, Limbach PA. RNAModMapper: RNA Modification Mapping Software for Analysis of Liquid Chromatography Tandem Mass Spectrometry Data. Anal Chem, 2017, 89(20): 10744-10752.
pmid: 28942636 |
| [126] |
Lobue PA, Yu NX, Jora M, Abernathy S, Limbach PA. Improved application of rnamodmapper - an RNA modification mapping software tool - for analysis of liquid chromatography tandem mass spectrometry (LC-MS/MS) data. Methods, 2019, 156: 128-138.
pmid: 30366097 |
| [127] |
Nakayama H, Akiyama M, Taoka M, Yamauchi Y, Nobe Y, Ishikawa H, Takahashi N, Isobe T. Ariadne: a database search engine for identification and chemical analysis of RNA using tandem mass spectrometry data. Nucleic Acids Res, 2009, 37(6): e47.
pmid: 19270066 |
| [128] |
Paulines MJ, Wetzel C, Limbach PA. Using spectral matching to interpret LC-MS/MS data during RNA modification mapping. J Mass Spectrom, 2019, 54(11): 906-914.
pmid: 31663233 |
| [129] |
Sample PJ, Gaston KW, Alfonzo JD, Limbach PA. RoboOligo: software for mass spectrometry data to support manual and de novo sequencing of post- transcriptionally modified ribonucleic acids. Nucleic Acids Res, 2015, 43(10): e64.
pmid: 25820423 |
| [130] |
Taoka M, Nobe Y, Hori M, Takeuchi A, Masaki S, Yamauchi Y, Nakayama H, Takahashi N, Isobe T. A mass spectrometry-based method for comprehensive quantitative determination of post-transcriptional RNA modifications: the complete chemical structure of Schizosaccharomyces pombe ribosomal RNAs. Nucleic Acids Res, 2015, 43(18): e115.
pmid: 26013808 |
| [131] |
Lanzillotti MB, Brodbelt JS. Nucleo-SAFARI: automated identification of fragment Ions in top-down MS/MS spectra of nucleic acids. Anal Chem, 2024, 96(41): 16115-16120.
pmid: 39365982 |
| [132] |
Cao XN, Zhang YY, Ding YL, Wan Y. Identification of RNA structures and their roles in RNA functions. Nat Rev Mol Cell Biol, 2024, 25(10): 784-801.
pmid: 38926530 |
| [133] |
Bonilla SL, Jones AN, Incarnato D. Structural and biophysical dissection of RNA conformational ensembles. Curr Opin Struct Biol, 2024, 88: 102908.
pmid: 39146886 |
| [134] |
Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J Am Chem Soc, 2005, 127(12): 4223-4231.
pmid: 15783204 |
| [135] |
Scalabrin M, Siu Y, Asare-Okai PN, Fabris D. Structure- specific ribonucleases for MS-based elucidation of higher-order RNA structure. J Am Soc Mass Spectrom, 2014, 25(7): 1136-1145.
pmid: 24845355 |
| [136] |
Sosic A, Göttlich R, Fabris D, Gatto B. B-CePs as cross-linking probes for the investigation of RNA higher-order structure. Nucleic Acids Res, 2021, 49(12): 6660-6672.
pmid: 34125908 |
| [137] |
Schneeberger EM, Halper M, Palasser M, Heel SV, Vušurović J, Plangger R, Juen M, Kreutz C, Breuker K. Native mass spectrometry reveals the initial binding events of HIV-1 rev to RRE stem II RNA. Nat Commun, 2020, 11(1): 5750.
pmid: 33188169 |
| [138] |
Kenderdine T, Fabris D. The multifaceted roles of mass spectrometric analysis in nucleic acids drug discovery and development. Mass Spectrom Rev, 2023, 42(4): 1332-1357.
pmid: 34939674 |
| [139] |
Karch KR, Snyder DT, Harvey SR, Wysocki VH. Native mass spectrometry: recent progress and remaining challenges. Annu Rev Biophys, 2022, 51: 157-179.
pmid: 34982572 |
| [140] |
Schneeberger EM, Breuker K. Native top-down mass spectrometry of tar RNA in complexes with a wild-type tat peptide for binding site mapping. Angew Chem Int Ed Engl, 2017, 56(5): 1254-1258.
pmid: 28000363 |
| [1] | 宋鹏辉, 马丽娟, 严冬. 外显子拼接复合体塑造m6A表观转录组的形成[J]. 遗传, 2023, 45(6): 464-471. |
| [2] | 王娟, 杨悦宁, 朴威兰, 金花. 尿苷酸化:一种重要的细胞内RNA监控方式[J]. 遗传, 2022, 44(6): 449-465. |
| [3] | 张婷婷, 刘峰. 斑马鱼蛋白酪氨酸硫酸化修饰的检测方法研究[J]. 遗传, 2022, 44(2): 178-186. |
| [4] | 张凤霞,王国栋. 现代代谢组学平台建设及相关技术应用[J]. 遗传, 2019, 41(9): 883-892. |
| [5] | 薛鹏, 蒋涛, 沈兴家. m 6A修饰及其对病毒复制过程调控研究进展[J]. 遗传, 2019, 41(5): 404-412. |
| [6] | 杨莹,陈宇晟,孙宝发,杨运桂. RNA甲基化修饰调控和规律[J]. 遗传, 2018, 40(11): 964-976. |
| [7] | 初芹, 李东, 侯诗宇, 石万海, 刘林, 王雅春. 基于DNA池测序法筛选奶牛高信息量SNP标记的可行性[J]. 遗传, 2014, 36(7): 691-696. |
| [8] | 李语丽 于军 宋述慧. RNA中6-甲基腺嘌呤的研究进展[J]. 遗传, 2013, 35(12): 1340-1351. |
| [9] | 李灏,姜颖,贺福初. 代谢组学技术及其在临床研究中的应用[J]. 遗传, 2008, 30(4): 389-399. |
| [10] | 邓新宇,姜颖,贺福初. 磷酸化蛋白质及多肽相关研究的技术进展[J]. 遗传, 2007, 29(10): 1163-1166. |
| [11] | 阮松林,马华升,王世恒,忻 雅,钱丽华,童建新,赵杭苹,王 杰. 植物蛋白质组学研究进展Ⅰ. 蛋白质组关键技术[J]. 遗传, 2006, 28(11): 1472-1486. |
| [12] | 吴松锋,朱云平,贺福初. 人类蛋白质组表达谱蛋白质鉴定的分步搜索策略[J]. 遗传, 2005, 27(5): 687-693. |
| [13] | 赵广荣,扬帆,元英进,髙秀梅,张军平. 单核苷酸多态性检测方法的新进展[J]. 遗传, 2005, 27(1): 123-129. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||

www.chinagene.cn
备案号:京ICP备09063187号-4
总访问:,今日访问:,当前在线:
