遗传 ›› 2024, Vol. 46 ›› Issue (9): 716-726.doi: 10.16288/j.yczz.24-124
杨森( ), 马宝霞, 钱泓润, 崔婕妤, 张潇筠, 李利达, 魏泽辉, 张智英, 王建刚(
), 马宝霞, 钱泓润, 崔婕妤, 张潇筠, 李利达, 魏泽辉, 张智英, 王建刚( ), 徐坤(
), 徐坤( )
)
收稿日期:2024-06-28
修回日期:2024-08-24
出版日期:2024-08-26
发布日期:2024-08-26
通讯作者:
王建刚,博士,讲师,研究方向:动物分子设计育种。E-mail: wangjiangang@126.com;作者简介:杨森,硕士研究生,专业方向:动物生物技术。E-mail: yangsen@nwafu.edu.cn
基金资助:
Sen Yang( ), Baoxia Ma, Hongrun Qian, Jieyu Cui, Xiaojun Zhang, Lida Li, Zehui Wei, Zhiying Zhang, Jiangang Wang(
), Baoxia Ma, Hongrun Qian, Jieyu Cui, Xiaojun Zhang, Lida Li, Zehui Wei, Zhiying Zhang, Jiangang Wang( ), Kun Xu(
), Kun Xu( )
)
Received:2024-06-28
Revised:2024-08-24
Published:2024-08-26
Online:2024-08-26
Supported by:摘要:
在哺乳动物细胞中,利用同源引导修复(homology-directed repair,HDR)机制的基因编辑策略能够实现精准的点编辑和敲入,但是HDR的低效性严重制约了该策略在精准医疗和分子设计育种中的应用。鉴于HDR机制所需的供体DNA模板不能自主募集到基因组双链断裂(double-stranded break,DSB)处,本课题组提出了供体适配系统(donor adapting system,DAS)的概念并开发出CRISPR/SpCas9-Gal4BD供体适配基因编辑系统。由于SpCas9蛋白分子较大,与Gal4BD适配器结合不利于表达、病毒载体包装及活体递送等过程,因此本研究进一步采用两种小型化Cas蛋白——路邓葡萄球菌(Staphylococcus lugdunensis)源SlugCas9变体SlugCas9-HF与氨基酸球菌(Acidaminococcus sp.)源AsCas12a,开发了新型的CRISPR/Gal4BD-SlugCas9和CRISPR/Gal4BD-AsCas12a供体适配基因编辑系统。通过SSA活性报告实验初步证明了Gal4BD与SlugCas9、AsCas12a N-端融合对其打靶活性影响较小。通过HDR效率报告实验进行功能验证并优化供体设计方案,结果表明:对于CRISPR/Gal4BD-AsCas12a DAS,适配器结合序列(binding sequence,BS)与供体5′-端融合(BS-dsDNA)效果较好;对于CRISPR/Gal4BD-SlugCas9 DAS,则BS与供体3′-端融合(dsDNA-BS)较佳。最终,利用CRISPR/Gal4BD-SlugCas9 DAS对HEK293T细胞中的EMX1、NUDT5、AAVS1三个基因位点分别实现了24%、37%、31%的精准编辑,相比对照组得到了显著提高。本研究为供体适配基因编辑系统的进一步优化提供了参考和借鉴,为后续动物分子设计育种应用研究提供了新的基因编辑工具。
杨森, 马宝霞, 钱泓润, 崔婕妤, 张潇筠, 李利达, 魏泽辉, 张智英, 王建刚, 徐坤. 基于小型化Cas蛋白的CRISPR/Gal4BD-Cas供体适配基因编辑系统研究[J]. 遗传, 2024, 46(9): 716-726.
Sen Yang, Baoxia Ma, Hongrun Qian, Jieyu Cui, Xiaojun Zhang, Lida Li, Zehui Wei, Zhiying Zhang, Jiangang Wang, Kun Xu. CRISPR/Gal4BD-Cas donor adapting systems based on miniaturized Cas proteins for improved gene editing[J]. Hereditas(Beijing), 2024, 46(9): 716-726.
表1
不同小型化Cas蛋白的比较"
| 蛋白名称 | 蛋白来源 | 类型 | 蛋白长度 | PAM序列 | gRNA类型 | 参考文献 |
|---|---|---|---|---|---|---|
| SaCas9 | Staphylococcus aureus | II-A | 1053 aa | -NNGRRT | sgRNA | [ |
| SchCas9 | Staphylococcus chromogenes | II-A | 1054 aa | -NNGR | tracrRNA | [ |
| SlugCas9 | Staphylococcus lugdunensis | II-A | 1054 aa | -NNGG | sgRNA | [ |
| CjCas9 | Campylobacter jejuni | II-C | 984 aa | -NNNNRYAC | sgRNA | [ |
| NmeCas9 | N. meningitidis strain 8013 | II-C | 1081 aa | -NNNNGATT | sgRNA | [ |
| AsCas12a | Acidaminococcus | V-A | 1307 aa | TTTV- | crRNA | [ |
| LbCas12a | Lachnospiraceae bacterium | V-A | 1228 aa | TTTV- | crRNA | [ |
| Cas12c | − | V-C | 1209~1330 aa | 5′TG/5′TN | tracrRNA | [ |
| Cas12j(Casφ) | Biggiephage | V-J | 700~800 aa | TTN- | crRNA | [ |
| AsCas12f1 | Acidibacillus sulfuroxidans | V-F | 422 aa | NTTR- | sgRNA | [ |
| SpCas12f1 | Syntrophomonas palmitatica | V-F | 497 aa | TTC- | sgRNA | [ |
| CasMINI | Engineering from Un1Cas12f1 | V-F | 529 aa | TTTR- | sgRNA | [ |
| Cas12h | − | V-H | 870~924 aa | 5′RTR | crRNA | [ |
| Cas12i | − | V-I | 1033~1093 aa | 5′TTN | crRNA | [ |
| Cas12g | − | V-G | 720~830 aa | 无 | crRNA、tracrRNA | [ |

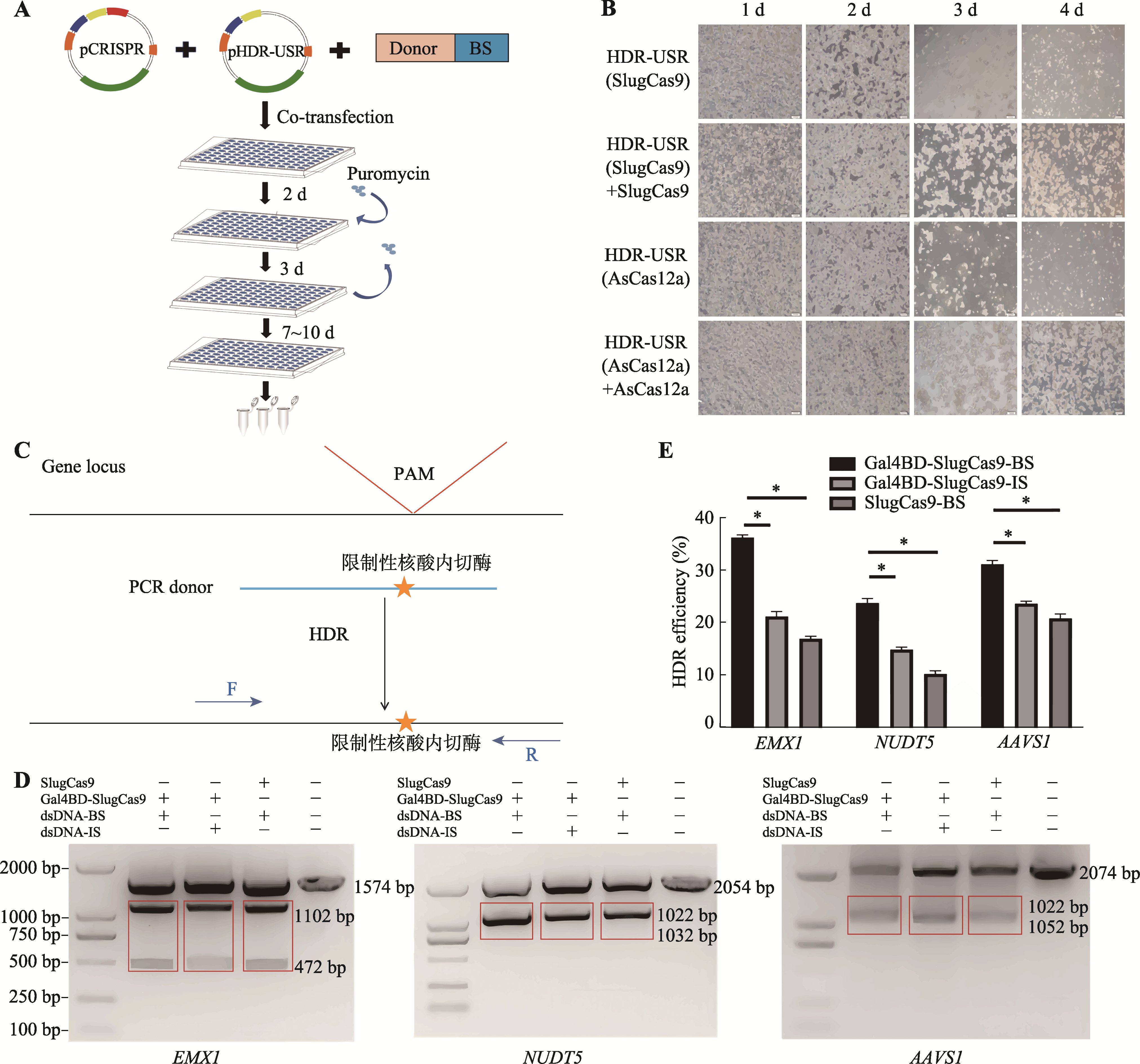
图4
CRISPR/Gal4BD-SlugCas9 DAS介导的点编辑 A:细胞基因组编辑与富集的流程图。B:pHDR-USR富集载体效果验证。共转染pHDR-USR载体与对应Cas蛋白作为实验组,单转染pHDR-USR载体作为对照组,转染48 h后加入puromycin进行筛选,连续4天固定时间在10倍镜下拍摄的细胞照片。C:基因位点基因编辑设计及检测示意图。其中一条PCR检测引物设计在dsDNA供体模板之外。D:RFLP实验检测SlugCas9介导HDR编辑效率的琼脂糖凝胶电泳结果。检测基因包括EMX1、NUDT5、AAVS1三个基因位点。E:基于RFLP 实验DNA条带灰度分析的HDR编辑效率检测结果。数据以平均值±SD表示,n=3;*:P<0.05。"
| [1] | Wu YX, Zeng J, Roscoe BP, Liu PP, Yao QM, Lazzarotto CR, Clement K, Cole MA, Luk K, Baricordi C, Shen AH, Ren CY, Esrick EB, Manis JP, Dorfman DM, Williams DA, Biffi A, Brugnara C, Biasco L, Brendel C, Pinello L, Tsai SQ, Wolfe SA, Bauer DE. Highly efficient therapeutic gene editing of human hematopoietic stem cells. Nat Med, 2019, 25(5): 776-783. |
| [2] |
Chen YO, Bao Y, Ma HZ, Yi ZY, Zhou Z, Wei WS. Gene editing technology and its research progress in China. Hereditas(Beijing), 2018, 40(10): 900-915.
doi: 10.16288/j.yczz.18-195 pmid: 30369472 |
| 陈一欧, 宝颖, 马华峥, 伊宗裔, 周卓, 魏文胜. 基因编辑技术及其在中国的研究发展. 遗传, 2018, 40(10): 900-915. | |
| [3] |
Nambiar TS, Baudrier L, Billon P, Ciccia A. CRISPR- based genome editing through the lens of DNA repair. Mol Cell, 2022, 82(2): 348-388.
doi: 10.1016/j.molcel.2021.12.026 pmid: 35063100 |
| [4] | Scully R, Panday A, Elango R, Willis NA. DNA double- strand break repair-pathway choice in somatic mammalian cells. Nat Rev Mol Cell Biol, 2019, 20(11): 698-714. |
| [5] | Paix A, Folkmann A, Goldman DH, Kulaga H, Grzelak MJ, Rasoloson D, Paidemarry S, Green R, Reed RR, Seydoux G. Precision genome editing using synthesis-dependent repair of Cas9-induced DNA breaks. Proc Natl Acad Sci USA, 2017, 114(50): E10745-E10754. |
| [6] |
Vasquez KM, Marburger K, Intody Z, Wilson JH. Manipulating the mammalian genome by homologous recombination. Proc Natl Acad Sci USA, 2001, 98(15): 8403-8410.
doi: 10.1073/pnas.111009698 pmid: 11459982 |
| [7] |
Nambiar TS, Billon P, Diedenhofen G, Hayward SB, Taglialatela A, Cai KH, Huang JW, Leuzzi G, Cuella-Martin R, Palacios A, Gupta A, Egli D, Ciccia A. Stimulation of CRISPR-mediated homology-directed repair by an engineered RAD18 variant. Nat Commun, 2019, 10(1): 3395.
doi: 10.1038/s41467-019-11105-z pmid: 31363085 |
| [8] |
Riesenberg S, Maricic T. Targeting repair pathways with small molecules increases precise genome editing in pluripotent stem cells. Nat Commun, 2018, 9(1): 2164.
doi: 10.1038/s41467-018-04609-7 pmid: 29867139 |
| [9] |
Wienert B, Nguyen DN, Guenther A, Feng SJ, Locke MN, Wyman SK, Shin J, Kazane KR, Gregory GL, Carter MAM, Wright F, Conklin BR, Marson A, Richardson CD, Corn JE. Timed inhibition of CDC7 increases CRISPR- Cas9 mediated templated repair. Nat Commun, 2020, 11(1): 2109.
doi: 10.1038/s41467-020-15845-1 pmid: 32355159 |
| [10] |
Ma M, Zhuang FF, Hu XB, Wang BL, Wen XZ, Ji JF, Jeff Xi J. Efficient generation of mice carrying homozygous double-floxp alleles using the Cas9-Avidin/Biotin-donor DNA system. Cell Res, 2017, 27(4): 578-581.
doi: 10.1038/cr.2017.29 pmid: 28266543 |
| [11] |
Carlson-Stevermer J, Abdeen AA, Kohlenberg L, Goedland M, Molugu K, Lou M, Saha K. Assembly of CRISPR ribonucleoproteins with biotinylated oligonucleotides via an RNA aptamer for precise gene editing. Nat Commun, 2017, 8(1): 1711.
doi: 10.1038/s41467-017-01875-9 pmid: 29167458 |
| [12] |
Ali Z, Shami A, Sedeek K, Kamel R, Alhabsi A, Tehseen M, Hassan N, Butt H, Kababji A, Hamdan SM, Mahfouz MM. Fusion of the Cas9 endonuclease and the VirD2 relaxase facilitates homology-directed repair for precise genome engineering in rice. Commun Biol, 2020, 3(1): 44.
doi: 10.1038/s42003-020-0768-9 pmid: 31974493 |
| [13] |
Keegan L, Gill G, Ptashne M. Separation of DNA binding from the transcription-activating function of a eukaryotic regulatory protein. Science, 1986, 231(4739): 699-704.
pmid: 3080805 |
| [14] | Giniger E, Ptashne M. Cooperative DNA binding of the yeast transcriptional activator GAL4. Proc Natl Acad Sci USA, 1988, 85(2): 382-386. |
| [15] | Zhang XJ, Xu K, Shen JC, Mu L, Qian HR, Cui JY, Ma BX, Chen ZL, Zhang ZY, Wei ZH. A CRISPR/Cas9-Gal4BD donor adapting system for enhancing homology-directed repair. Hereditas(Beijing), 2022, 44(8): 708-719. |
| 张潇筠, 徐坤, 沈俊岑, 穆璐, 钱泓润, 崔婕妤, 马宝霞, 陈知龙, 张智英, 魏泽辉. 一种新型提高HDR效率的CRISPR/Cas9-Gal4BD供体适配基因编辑系统. 遗传, 2022, 44(8): 708-719. | |
| [16] |
Kleinstiver BP, Sousa AA, Walton RT, Tak YE, Hsu JY, Clement K, Welch MM, Horng JE, Malagon-Lopez J, Scarfò I, Maus MV, Pinello L, Aryee MJ, Joung JK. Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat Biotechnol, 2019, 37(3): 276-282.
doi: 10.1038/s41587-018-0011-0 pmid: 30742127 |
| [17] |
Tóth E, Varga É, Kulcsár PI, Kocsis-Jutka V, Krausz SL, Nyeste A, Welker Z, Huszár K, Ligeti Z, Tálas A, Welker E. Improved LbCas12a variants with altered PAM specificities further broaden the genome targeting range of Cas12a nucleases. Nucleic Acids Res, 2020, 48(7): 3722-3733.
doi: 10.1093/nar/gkaa110 pmid: 32107556 |
| [18] | Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu XB, Makarova KS, Koonin EV, Sharp PA, Zhang F. In vivo genome editing using Staphylococcus aureus Cas9. Nature, 2015, 520(7546): 186-191. |
| [19] | Wang S, Mao HL, Hou LH, Hu ZY, Wang Y, Qi T, Tao C, Yang Y, Zhang CD, Li MM, Liu HH, Hu SJ, Chai RJ, Wang YM. Compact SchCas9 recognizes the simple NNGR PAM. Adv Sci (Weinh), 2022, 9(4): e2104789. |
| [20] |
Hu ZY, Zhang CD, Wang S, Gao SQ, Wei JJ, Li MM, Hou LH, Mao HL, Wei YY, Qi T, Liu HM, Liu D, Lan F, Lu DR, Wang HY, Li JX, Wang YM. Discovery and engineering of small SlugCas9 with broad targeting range and high specificity and activity. Nucleic Acids Res, 2021, 49(7): 4008-4019.
doi: 10.1093/nar/gkab148 pmid: 33721016 |
| [21] | Kim E, Koo T, Park SW, Kim D, Kim K, Cho HY, Song DW, Lee KJ, Jung MH, Kim S, Kim JH, Kim JH, Kim JS. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat Commun, 2017, 8: 14500. |
| [22] | Hou ZG, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer EJ, Thomson JA. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci USA, 2013, 110(39): 15644-15649. |
| [23] |
Pausch P, Al-Shayeb B, Bisom-Rapp E, Tsuchida CA, Li Z, Cress BF, Knott GJ, Jacobsen SE, Banfield JF, Doudna JA. CRISPR-CasΦ from huge phages is a hypercompact genome editor. Science, 2020, 369(6501): 333-337.
doi: 10.1126/science.abb1400 pmid: 32675376 |
| [24] |
Wu ZW, Zhang YF, Yu HP, Pan D, Wang YJ, Wang YN, Li F, Liu C, Nan H, Chen WZ, Ji QJ. Programmed genome editing by a miniature CRISPR-Cas12f nuclease. Nat Chem Biol, 2021, 17(11): 1132-1138.
doi: 10.1038/s41589-021-00868-6 pmid: 34475565 |
| [25] |
Bigelyte G, Young JK, Karvelis T, Budre K, Zedaveinyte R, Djukanovic V, Van Ginkel E, Paulraj S, Gasior S, Jones S, Feigenbutz L, Clair GS, Barone P, Bohn J, Acharya A, Zastrow-Hayes G, Henkel-Heinecke S, Silanskas A, Seidel R, Siksnys V. Miniature type V-F CRISPR-Cas nucleases enable targeted DNA modification in cells. Nat Commun, 2021, 12(1): 6191.
doi: 10.1038/s41467-021-26469-4 pmid: 34702830 |
| [26] | Kim DY, Lee JM, Moon SB, Chin HJ, Park S, Lim Y, Kim D, Koo T, Ko JH, Kim YS. Efficient CRISPR editing with a hypercompact Cas12f1 and engineered guide RNAs delivered by adeno-associated virus. Nat Biotechnol, 2022, 40(1): 94-102. |
| [27] |
Yan WX, Hunnewell P, Alfonse LE, Carte JM, Keston- Smith E, Sothiselvam S, Garrity AJ, Chong SR, Makarova KS, Koonin EV, Cheng DR, Scott DA. Functionally diverse type V CRISPR-Cas systems. Science, 2019, 363(6422): 88-91.
doi: 10.1126/science.aav7271 pmid: 30523077 |
| [28] |
Zhang LY, Zuris JA, Viswanathan R, Edelstein JN, Turk R, Thommandru B, Rube HT, Glenn SE, Collingwood MA, Bode NM, Beaudoin SF, Lele S, Scott SN, Wasko KM, Sexton S, Borges CM, Schubert MS, Kurgan GL, McNeill MS, Fernandez CA, Myer VE, Morgan RA, Behlke MA, Vakulskas CA. AsCas12a ultra nuclease facilitates the rapid generation of therapeutic cell medicines. Nat Commun, 2021, 12(1): 3908.
doi: 10.1038/s41467-021-24017-8 pmid: 34162850 |
| [29] |
Fagerlund RD, Staals RHJ, Fineran PC. The Cpf1 CRISPR-Cas protein expands genome-editing tools. Genome Biol, 2015, 16: 251.
doi: 10.1186/s13059-015-0824-9 pmid: 26578176 |
| [30] | Shao SM, Ren CH, Liu ZT, Bai YC, Chen ZL, Wei ZH, Wang X, Zhang ZY, Xu K. Enhancing CRISPR/ Cas9-mediated homology-directed repair in mammalian cells by expressing Saccharomyces cerevisiae Rad52. Int J Biochem Cell Biol, 2017, 92: 43-52. |
| [31] | Xu K, Ren CH, Liu ZT, Zhang T, Zhang TT, Li D, Wang L, Yan Q, Guo LJ, Shen JC, Zhang ZY. Efficient genome engineering in eukaryotes using Cas9 from Streptococcus thermophilus. Cell Mol Life Sci, 2015, 72(2): 383-399. |
| [32] | Yan NN, Sun YS, Fang YY, Deng JR, Mu L, Xu K, Mymryk JS, Zhang ZY. A universal surrogate reporter for efficient enrichment of CRISPR/Cas9-mediated homology- directed repair in mammalian cells. Mol Ther Nucleic Acids, 2020, 19: 775-789. |
| [33] | Qian WD, Huang J, Wang XF, Wang T, Li YD. CRISPR-Cas12a combined with reverse transcription recombinase polymerase amplification for sensitive and specific detection of human norovirus genotype GII.4. Virology, 2021, 564: 26-32. |
| [34] | Zheng SY, Ma LL, Wang XL, Lu LX, Ma ST, Xu B, Ouyang W. RPA-Cas12aDS: A visual and fast molecular diagnostics platform based on RPA-CRISPR-Cas12a method for infectious bursal disease virus detection. J Virol Methods, 2022, 304: 114523. |
| [35] |
Nguyen DN, Roth TL, Li PJ, Chen PA, Apathy R, Mamedov MR, Vo LT, Tobin VR, Goodman D, Shifrut E, Bluestone JA, Puck JM, Szoka FC, Marson A. Polymer-stabilized Cas9 nanoparticles and modified repair templates increase genome editing efficiency. Nat Biotechnol, 2020, 38(1): 44-49.
doi: 10.1038/s41587-019-0325-6 pmid: 31819258 |
| [36] | Savic N, Ringnalda FC, Lindsay H, Berk C, Bargsten K, Li YZ, Neri D, Robinson MD, Ciaudo C, Hall J, Jinek M, Schwank G. Covalent linkage of the DNA repair template to the CRISPR-Cas9 nuclease enhances homology- directed repair. eLife, 2018, 7: e33761. |
| [37] |
Ma SF, Wang XL, Hu YF, Lv J, Liu CF, Liao KT, Guo XH, Wang D, Lin Y, Rong ZL. Enhancing site-specific DNA integration by a Cas9 nuclease fused with a DNA donor-binding domain. Nucleic Acids Res, 2020, 48(18): 10590-10601.
doi: 10.1093/nar/gkaa779 pmid: 32986839 |
| [1] | 潘东霞, 王辉, 熊本海, 唐湘方. CRISPR-Cas基因编辑技术在羊生产应用中研究进展[J]. 遗传, 2024, 46(9): 690-700. |
| [2] | 马宝霞, 杨森, 吕明, 王昱人, 常立业, 韩艺帆, 王建刚, 郭杨, 徐坤. 不同CRISPR/Cas9供体适配基因编辑系统的比较及优化研究[J]. 遗传, 2024, 46(6): 466-477. |
| [3] | 曹振林, 李金红, 周铭辉, 张曼婷, 王宁, 陈一飞, 李嘉欣, 祝青松, 宫雯珺, 杨绪晨, 方小龙, 和家贤, 李美娜. 大豆花药优势表达基因GmFLA22a调控雄性育性的功能研究[J]. 遗传, 2024, 46(4): 333-345. |
| [4] | 鲍艳春, 戴伶俐, 刘在霞, 马凤英, 王宇, 刘永斌, 谷明娟, 娜日苏, 张文广. CRISPR/Cas9系统在畜禽遗传改良中研究进展[J]. 遗传, 2024, 46(3): 219-231. |
| [5] | 卞中, 曹东平, 庄文姝, 张舒玮, 刘巧泉, 张林. 水稻分子设计育种启示:传统与现代相结合[J]. 遗传, 2023, 45(9): 718-740. |
| [6] | 王秉政, 张超, 张佳丽, 孙锦. 利用单转录本表达Cas9和sgRNA条件性编辑果蝇基因组[J]. 遗传, 2023, 45(7): 593-601. |
| [7] | 吴仲胜, 高誉, 杜勇涛, 党颂, 何康敏. CRISPR-Cas9基因编辑技术对细胞内源蛋白进行荧光标记的实验操作[J]. 遗传, 2023, 45(2): 165-175. |
| [8] | 刘梅珍, 王立人, 李咏梅, 马雪云, 韩红辉, 李大力. 利用CRISPR/Cas9技术构建基因编辑大鼠模型[J]. 遗传, 2023, 45(1): 78-87. |
| [9] | 张潇筠, 徐坤, 沈俊岑, 穆璐, 钱泓润, 崔婕妤, 马宝霞, 陈知龙, 张智英, 魏泽辉. 一种新型提高HDR效率的CRISPR/Cas9-Gal4BD供体适配基因编辑系统[J]. 遗传, 2022, 44(8): 708-719. |
| [10] | 韩玉婷, 许博文, 李羽童, 卢心怡, 董习之, 邱雨浩, 车沁耘, 朱芮葆, 郑丽, 李孝宸, 司绪, 倪建泉. 模式动物果蝇的基因调控前沿技术[J]. 遗传, 2022, 44(1): 3-14. |
| [11] | 王海涛, 李亭亭, 黄勋, 马润林, 刘秋月. 遗传修饰技术在绵羊分子设计育种中的应用[J]. 遗传, 2021, 43(6): 580-600. |
| [12] | 彭定威, 李瑞强, 曾武, 王敏, 石翾, 曾检华, 刘小红, 陈瑶生, 何祖勇. 编辑MSTN半胱氨酸节基元促进两广小花猪肌肉生长[J]. 遗传, 2021, 43(3): 261-270. |
| [13] | 李国玲, 杨善欣, 吴珍芳, 张献伟. 提高CRISPR/Cas9介导的动物基因组精确插入效率 研究进展[J]. 遗传, 2020, 42(7): 641-656. |
| [14] | 陈赢男, 陆静. CRISPR/Cas9系统在林木基因编辑中的应用[J]. 遗传, 2020, 42(7): 657-668. |
| [15] | 李霞, 施皖, 耿立召, 许建平. CRISPR/Cas核糖核蛋白介导的植物基因组编辑[J]. 遗传, 2020, 42(6): 556-564. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||

www.chinagene.cn
备案号:京ICP备09063187号-4
总访问:,今日访问:,当前在线:
