遗传 ›› 2025, Vol. 47 ›› Issue (1): 34-45.doi: 10.16288/j.yczz.24-196
收稿日期:2024-07-01
修回日期:2024-10-08
出版日期:2025-01-20
发布日期:2024-11-06
通讯作者:
崔银秋,博士,教授,研究方向:古基因组学。E-mail: cuiyq@jlu.edu.cn作者简介:张达轩,博士研究生,专业方向:生物化学与分子生物学。E-mail: dxzhang20@mails.jlu.edu.cn
基金资助:
Daxuan Zhang( ), Shenru Dai, Yinqiu Cui(
), Shenru Dai, Yinqiu Cui( )
)
Received:2024-07-01
Revised:2024-10-08
Published:2025-01-20
Online:2024-11-06
Supported by:摘要:
广袤的亚洲北部地区,包括西伯利亚、蒙古高原以及中国北方地区,不仅是欧亚大陆上人群交流的十字路口,也是连接美洲大陆的重要桥梁,这一区域对于探索人类起源、追踪人类迁移路线以及阐释演化机制具有独特且不可替代的重要性。尽管由于样本发掘数量有限、保存状态不一和技术手段的制约,人们对亚洲北部地区人群互动过程的理解尚处于初步阶段,但高通量测序技术的发展及其在古DNA研究中的进展为人们提供了从分子层面深入探究古代人群遗传历史的全新视角。本文通过综合分析亚洲北部地区不同阶段古代人群的遗传结构变化,旨在揭示该地区古代人群间的互动模式、遗传结构的演变过程以及他们对现代人群的遗传贡献;此外,本文还探讨了人类应对极端自然条件的适应性策略。这些研究不仅加深了人们对人类起源和迁移过程的理解,还将为研究人类在环境选择压力下的演化机制和适应性策略提供了坚实的基础。
张达轩, 戴沈汝, 崔银秋. 古基因组视角下的亚洲北部人群迁徙和演化机制[J]. 遗传, 2025, 47(1): 34-45.
Daxuan Zhang, Shenru Dai, Yinqiu Cui. The migration and evolutionary mechanisms of northern Asian populations from the perspective of ancient genomics[J]. Hereditas(Beijing), 2025, 47(1): 34-45.
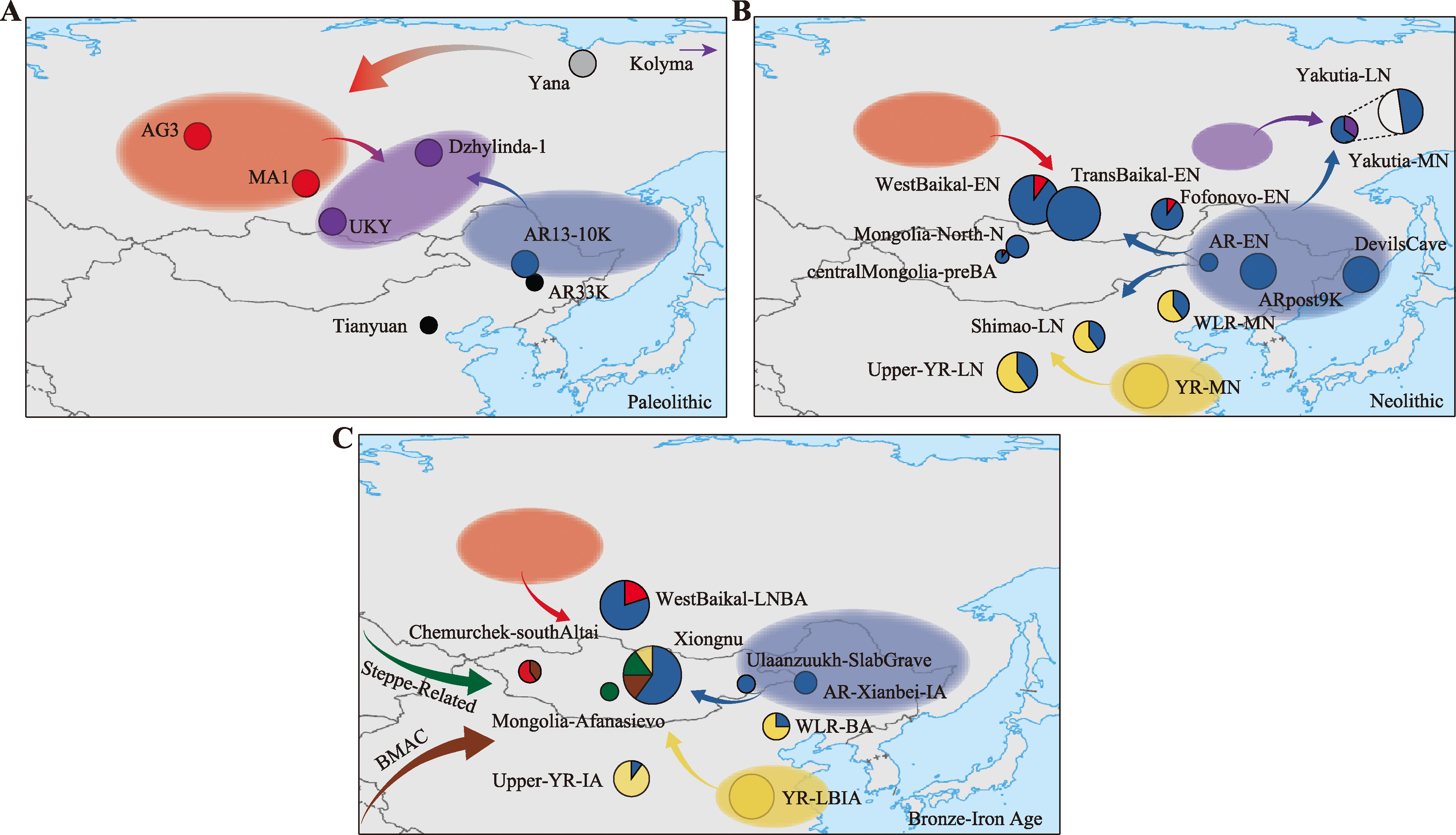
图1
古代人群的地理分布图及遗传结构示意图 A:旧石器时代(更新世晚期-全新世)。Tianyuan和AR33K为亚洲早期古代人群遗传结构主要代表;Yana为ANS遗传结构的主要代表;MA1和AG3为ANE遗传结构的主要代表;AR13-10K为ANA遗传结构的主要代表,UKY、Dzhylinda-1和Kolyma为APS遗传结构的主要代表。ANE遗传结构与ANS遗传结构具有紧密联系,是ANS遗传结构的后裔;ANE遗传结构与ANA遗传结构还有美洲土著人群遗传结构共同形成了独特的APS遗传结构。B:新石器时代。贝加尔湖地区WestBaikal-EN、Fofonovo-EN与蒙古高原地区centralMongolia-preBA人群遗传结构是ANA遗传结构与ANE遗传结构之间交流互动的结果,同时在贝加尔湖地区TransBaikal-EN人群与蒙古地区Mongolia-North-N人群都属于ANA遗传结构;俄罗斯雅库特地区Yakutia-MN人群遗传结构是ANA遗传结构与APS遗传结构交流互动的结果,同时在该地区存在的遗传连续性,稍晚时期的Yakutia-LN人群遗传结构是Yakutia-MN人群遗传结构与ANA遗传结构交流互动的结果;黑龙江境内的AR-EN、ARpost9K人群与俄罗斯远东地区的DevilsCave人群与ANA遗传结构一致;黄河流域下游YR-MN人群是稳定且独特的黄河流域遗传结构的主要代表,黄河中上游Upper-YR-LN、Shimao-LN人群以及西辽河流域WLR-MN人群是YR-MN遗传结构与ANA遗传结构交流互动的结果。C:金属时代(包括青铜时代和铁器时代)。贝加尔湖地区WestBaikal-LNBA人群是ANA与ANE的遗传交流的结果;蒙古地区存在与青铜时代早期草原Afanasievo人群遗传结构完全一致的Mongolia-Afanasievo人群、与ANA遗传结构完全一致的Ulaanzuukh-SlabGrave人群、中亚(BMAC)人群遗传结构与ANE遗传结构交流互动产生的Chemurchek-southAltai人群以及与周边地区频繁互动产生的遗传结构高度异质化的Xiongnu人群;黑龙江境内存在文化上属于鲜卑人群,遗传结构上与ANA遗传结构完全一致的AR-Xianbei-IA人群;黄河流域下游YR-LBIA人群保持着黄河流域遗传结构稳定性;黄河上游Upper-YR-IA人群以及西辽河流域WLR-BA人群是YR-MN遗传结构与ANA遗传结构交流互动的结果,分别与本地区较早人群相比具有更多黄河流域人群遗传成分。地图来自网站http://bzdt.ch.mnr.gov.cn/index.html,标准地图审图号GS(2016)2962。"
| [1] |
Yang MA, Gao X, Theunert C, Tong HW, Aximu-Petri A, Nickel B, Slatkin M, Meyer M, Pääbo S, Kelso J, Fu QM. 40,000-year-old individual from Asia provides insight into early population structure in Eurasia. Curr Biol, 2017, 27(20): 3202-3208.e9.
doi: S0960-9822(17)31195-8 pmid: 29033327 |
| [2] | Hajdinjak M, Mafessoni F, Skov L, Vernot B, Hübner A, Fu QM, Essel E, Nagel S, Nickel B, Richter J, Moldovan OT, Constantin S, Endarova E, Zahariev N, Spasov R, Welker F, Smith GM, Sinet-Mathiot V, Paskulin L, Fewlass H, Talamo S, Rezek Z, Sirakova S, Sirakov N, McPherron SP, Tsanova T, Hublin JJ, Peter BM, Meyer M, Skoglund P, Kelso J, Pääbo S. Initial Upper Palaeolithic humans in Europe had recent neanderthal ancestry. Nature, 2021, 592(7853): 253-257. |
| [3] | Sikora M, Pitulko VV, Sousa VC, Allentoft ME, Vinner L, Rasmussen S, Margaryan A, de Barros Damgaard P, de la Fuente C, Renaud G, Yang MA, Fu QM, Dupanloup I, Giampoudakis K, Nogués-Bravo D, Rahbek C, Kroonen G, Peyrot M, McColl H, Vasilyev SV, Veselovskaya E, Gerasimova M, Pavlova EY, Chasnyk VG, Nikolskiy PA, Gromov AV, Khartanovich VI, Moiseyev V, Grebenyuk PS, Fedorchenko AY, Lebedintsev AI, Slobodin SB, Malyarchuk BA, Martiniano R, Meldgaard M, Arppe L, Palo JU, Sundell T, Mannermaa K, Putkonen M, Alexandersen V, Primeau C, Baimukhanov N, Malhi RS, Sjögren KG, Kristiansen K, Wessman A, Sajantila A, Lahr MM, Durbin R, Nielsen R, Meltzer DJ, Excoffier L, Willerslev E. The population history of northeastern Siberia since the Pleistocene. Nature, 2019, 570(7760): 182-188. |
| [4] | Raghavan M, Skoglund P, Graf KE, Metspalu M, Albrechtsen A, Moltke I, Rasmussen S, Stafford TW, Orlando L, Metspalu E, Karmin M, Tambets K, Rootsi S, Mägi R, Campos PF, Balanovska E, Balanovsky O, Khusnutdinova E, Litvinov S, Osipova LP, Fedorova SA, Voevoda MI, DeGiorgio M, Sicheritz-Ponten T, Brunak S, Demeshchenko S, Kivisild T, Villems R, Nielsen R, Jakobsson M, Willerslev E. Upper Palaeolithic Siberian genome reveals dual ancestry of native Americans. Nature, 2014, 505(7481): 87-91. |
| [5] |
Mao XW, Zhang HC, Qiao SY, Liu YC, Chang FQ, Xie P, Zhang M, Wang TY, Li M, Cao P, Yang RW, Liu F, Dai QY, Feng XT, Ping WJ, Lei CZ, Olsen JW, Bennett EA, Fu QM. The deep population history of northern East Asia from the Late Pleistocene to the Holocene. Cell, 2021, 184(12): 3256-3266.e13.
doi: 10.1016/j.cell.2021.04.040 pmid: 34048699 |
| [6] | Osman MB, Tierney JE, Zhu J, Tardif R, Hakim GJ, King J, Poulsen CJ. Globally resolved surface temperatures since the Last Glacial Maximum. Nature, 2021, 599(7884): 239-244. |
| [7] | Ji DX, Chen FH, Bettinger RL, Elston RG, Geng ZQ, Barton L, Wang H, An CB, Zhang DG. Human response to the Last Glacial Maximum: evidence from North China. Acta Anthropologica Sinica, 2005, 24(4): 270-282. |
| 吉笃学, 陈发虎, Bettinger RL, Elston RG, 耿志强, Barton L, 王辉, 安成邦, 张东菊. 末次盛冰期环境恶化对中国北方旧石器文化的影响. 人类学学报, 2005, 24(4): 270-282. | |
| [8] | Fu QM, Posth C, Hajdinjak M, Petr M, Mallick S, Fernandes D, Furtwängler A, Haak W, Meyer M, Mittnik A, Nickel B, Peltzer A, Rohland N, Slon V, Talamo S, Lazaridis I, Lipson M, Mathieson I, Schiffels S, Skoglund P, Derevianko AP, Drozdov N, Slavinsky V, Tsybankov A, Cremonesi RG, Mallegni F, Gély B, Vacca E, Morales MRG, Straus LG, Neugebauer-Maresch C, Teschler- Nicola M, Constantin S, Moldovan OT, Benazzi S, Peresani M, Coppola D, Lari M, Ricci S, Ronchitelli A, Valentin F, Thevenet C, Wehrberger K, Grigorescu D, Rougier H, Crevecoeur I, Flas D, Semal P, Mannino MA, Cupillard C, Bocherens H, Conard NJ, Harvati K, Moiseyev V, Drucker DG, Svoboda J, Richards MP, Caramelli D, Pinhasi R, Kelso J, Patterson N, Krause J, Pääbo S, Reich D. The genetic history of Ice Age Europe. Nature, 2016, 534(7606): 200-205. |
| [9] |
Yu H, Spyrou MA, Karapetian M, Shnaider S, Radzevičiūtė R, Nägele K, Neumann GU, Penske S, Zech J, Lucas M, LeRoux P, Roberts P, Pavlenok G, Buzhilova A, Posth C, Jeong C, Krause J. Paleolithic to Bronze Age Siberians reveal connections with first Americans and across Eurasia. Cell, 2020, 181(6): 1232-1245.e20.
doi: S0092-8674(20)30502-X pmid: 32437661 |
| [10] | Kılınç GM, Kashuba N, Koptekin D, Bergfeldt N, Dönertaş HM, Rodríguez-Varela R, Shergin D, Ivanov G, Kichigin D, Pestereva K, Volkov D, Mandryka P, Kharinskii A, Tishkin A, Ineshin E, Kovychev E, Stepanov A, Dalén L, Günther T, Kırdök E, Jakobsson M, Somel M, Krzewińska M, Storå J, Götherström A. Human population dynamics and Yersinia pestis in ancient northeast Asia. Sci Adv, 2021, 7(2): eabc4587. |
| [11] | Gill H, Lee J, Jeong C. Reconstructing the genetic relationship between ancient and present-day Siberian populations. Genome Biol Evol, 2024, 16(4): evae063. |
| [12] |
Tallavaara M, Luoto M, Korhonen N, Järvinen H, Seppä H. Human population dynamics in Europe over the Last Glacial Maximum. Proc Natl Acad Sci USA, 2015, 112(27): 8232-8237.
doi: 10.1073/pnas.1503784112 pmid: 26100880 |
| [13] | Qian YP. A brief discussion on the cultural characteristics and starting marks of the Neolithic Age. West Archaeol, 2012, (1): 52-62. |
| 钱耀鹏. 略论新石器时代的文化特征与起始标志. 西部考古, 2012, (1): 52-62. | |
| [14] |
Lu HY, Zhang JP, Liu KB, Wu NQ, Li YM, Zhou KS, Ye ML, Zhang TY, Zhang HJ, Yang XY, Shen LC, Xu DK, Li Q. Earliest domestication of common millet (Panicum miliaceum) in East Asia extended to 10,000 years ago. Proc Natl Acad Sci USA, 2009, 106(18): 7367-7372.
doi: 10.1073/pnas.0900158106 pmid: 19383791 |
| [15] |
Liu XY, Jones MK, Zhao ZJ, Liu GX, O'Connell TC. The earliest evidence of millet as a staple crop: new light on Neolithic foodways in North China. Am J Phys Anthropol, 2012, 149(2): 283-290.
doi: 10.1002/ajpa.22127 pmid: 22961368 |
| [16] |
Ning C, Li TJ, Wang K, Zhang F, Li T, Wu XY, Gao SZ, Zhang QC, Zhang H, Hudson MJ, Dong GH, Wu SH, Fang YM, Liu C, Feng CY, Li W, Han T, Li R, Wei J, Zhu YG, Zhou YW, Wang CC, Fan SY, Xiong ZL, Sun ZY, Ye ML, Sun L, Wu XH, Liang FW, Cao YP, Wei XT, Zhu H, Zhou H, Krause J, Robbeets M, Jeong C, Cui YQ. Ancient genomes from northern China suggest links between subsistence changes and human migration. Nat Commun, 2020, 11(1): 2700.
doi: 10.1038/s41467-020-16557-2 pmid: 32483115 |
| [17] | Siska V, Jones ER, Jeon S, Bhak Y, Kim HM, Cho YS, Kim H, Lee K, Veselovskaya E, Balueva T, Gallego- Llorente M, Hofreiter M, Bradley DG, Eriksson A, Pinhasi R, Bhak J, Manica A. Genome-wide data from two early Neolithic East Asian individuals dating to 7700 years ago. Sci Adv, 2017, 3(2): e1601877. |
| [18] | Wang CC, Yeh HY, Popov AN, Zhang HQ, Matsumura H, Sirak K, Cheronet O, Kovalev A, Rohland N, Kim AM, Mallick S, Bernardos R, Tumen D, Zhao J, Liu YC, Liu JY, Mah M, Wang K, Zhang Z, Adamski N, Broomandkhoshbacht N, Callan K, Candilio F, Carlson KSD, Culleton BJ, Eccles L, Freilich S, Keating D, Lawson AM, Mandl K, Michel M, Oppenheimer J, Özdoğan KT, Stewardson K, Wen SQ, Yan S, Zalzala F, Chuang R, Huang CJ, Looh H, Shiung CC, Nikitin YG, Tabarev AV, Tishkin AA, Lin S, Sun ZY, Wu XM, Yang TL, Hu X, Chen L, Du H, Bayarsaikhan J, Mijiddorj E, Erdenebaatar D, Iderkhangai TO, Myagmar E, Kanzawa-Kiriyama H, Nishino M, Shinoda KI, Shubina OA, Guo JX, Cai WW, Deng QY, Kang LL, Li DW, Li DN, Lin R, Nini, Shrestha R, Wang LX, Wei LH, Xie GM, Yao HB, Zhang MF, He GL, Yang XM, Hu R, Robbeets M, Schiffels S, Kennett DJ, Jin L, Li H, Krause J, Pinhasi R, Reich D. Genomic insights into the formation of human populations in East Asia. Nature, 2021, 591(7850): 413-419. |
| [19] |
Jeong C, Wang K, Wilkin S, Taylor WTT, Miller BK, Bemmann JH, Stahl R, Chiovelli C, Knolle F, Ulziibayar S, Khatanbaatar D, Erdenebaatar D, Erdenebat U, Ochir A, Ankhsanaa G, Vanchigdash C, Ochir B, Munkhbayar C, Tumen D, Kovalev A, Kradin N, Bazarov BA, Miyagashev DA, Konovalov PB, Zhambaltarova E, Miller AV, Haak W, Schiffels S, Krause J, Boivin N, Erdene M, Hendy J, Warinner C. A dynamic 6,000-year genetic history of Eurasia's eastern steppe. Cell, 2020, 183(4): 890-904.e29.
doi: 10.1016/j.cell.2020.10.015 pmid: 33157037 |
| [20] |
Yang MA, Fan XC, Sun B, Chen CY, Lang JF, Ko YC, Tsang CH, Chiu H, Wang TY, Bao QC, Wu XH, Hajdinjak M, Ko AMS, Ding MY, Cao P, Yang RW, Liu F, Nickel B, Dai QY, Feng XT, Zhang LZ, Sun CK, Ning C, Zeng W, Zhao YS, Zhang M, Gao X, Cui YQ, Reich D, Stoneking M, Fu QM. Ancient DNA indicates human population shifts and admixture in northern and southern China. Science, 2020, 369(6501): 282-288.
doi: 10.1126/science.aba0909 pmid: 32409524 |
| [21] | de Barros Damgaard P, Martiniano R, Kamm J, Moreno- Mayar JV, Kroonen G, Peyrot M, Barjamovic G, Rasmussen S, Zacho C, Baimukhanov N, Zaibert V, Merz V, Biddanda A, Merz I, Loman V, Evdokimov V, Usmanova E, Hemphill B, Seguin-Orlando A, Yediay FE, Ullah I, Sjögren KG, Iversen KH, Choin J, de la Fuente C, Ilardo M, Schroeder H, Moiseyev V, Gromov A, Polyakov A, Omura S, Senyurt SY, Ahmad H, McKenzie C, Margaryan A, Hameed A, Samad A, Gul N, Khokhar MH, Goriunova OI, Bazaliiskii VI, Novembre J, Weber AW, Orlando L, Allentoft ME, Nielsen R, Kristiansen K, Sikora M, Outram AK, Durbin R, Willerslev E. The first horse herders and the impact of early Bronze Age steppe expansions into Asia. Science, 2018, 360(6396): eaar7711. |
| [22] | Jia X, Sun YG, Wang L, Sun WF, Zhao ZJ, Lee HF, Huang WB, Wu SY, Lu HY. The transition of human subsistence strategies in relation to climate change during the Bronze Age in the West Liao River Basin, northeast China. The Holocene, 2016, 26(5): 781-789. |
| [23] | Narasimhan VM, Patterson N, Moorjani P, Rohland N, Bernardos R, Mallick S, Lazaridis I, Nakatsuka N, Olalde I, Lipson M, Kim AM, Olivieri LM, Coppa A, Vidale M, Mallory J, Moiseyev V, Kitov E, Monge J, Adamski N, Alex N, Broomandkhoshbacht N, Candilio F, Callan K, Cheronet O, Culleton BJ, Ferry M, Fernandes D, Freilich S, Gamarra B, Gaudio D, Hajdinjak M, Harney É, Harper TK, Keating D, Lawson AM, Mah M, Mandl K, Michel M, Novak M, Oppenheimer J, Rai N, Sirak K, Slon V, Stewardson K, Zalzala F, Zhang Z, Akhatov G, Bagashev AN, Bagnera A, Baitanayev B, Bendezu-Sarmiento J, Bissembaev AA, Bonora GL, Chargynov TT, Chikisheva T, Dashkovskiy PK, Derevianko A, Dobeš M, Douka K, Dubova N, Duisengali MN, Enshin D, Epimakhov A, Fribus AV, Fuller D, Goryachev A, Gromov A, Grushin SP, Hanks B, Judd M, Kazizov E, Khokhlov A, Krygin AP, Kupriyanova E, Kuznetsov P, Luiselli D, Maksudov F, Mamedov AM, Mamirov TB, Meiklejohn C, Merrett DC, Micheli R, Mochalov O, Mustafokulov S, Nayak A, Pettener D, Potts R, Razhev D, Rykun M, Sarno S, Savenkova TM, Sikhymbaeva K, Slepchenko SM, Soltobaev OA, Stepanova N, Svyatko S, Tabaldiev K, Teschler-Nicola M, Tishkin AA, Tkachev VV, Vasilyev S, Velemínský P, Voyakin D, Yermolayeva A, Zahir M, Zubkov VS, Zubova A, Shinde VS, Lalueza-Fox C, Meyer M, Anthony D, Boivin N, Thangaraj K, Kennett DJ, Frachetti M, Pinhasi R, Reich D. The formation of human populations in South and Central Asia. Science, 2019, 365(6457): eaat7487. |
| [24] | Lee J, Miller BK, Bayarsaikhan J, Johannesson E, Ventresca Miller A, Warinner C, Jeong C. Genetic population structure of the Xiongnu Empire at imperial and local scales. Sci Adv, 2023, 9(15): eadf3904. |
| [25] | de Barros Damgaard P, Marchi N, Rasmussen S, Peyrot M, Renaud G, Korneliussen T, Moreno-Mayar JV, Pedersen MW, Goldberg A, Usmanova E, Baimukhanov N, Loman V, Hedeager L, Pedersen AG, Nielsen K, Afanasiev G, Akmatov K, Aldashev A, Alpaslan A, Baimbetov G, Bazaliiskii VI, Beisenov A, Boldbaatar B, Boldgiv B, Dorzhu C, Ellingvag S, Erdenebaatar D, Dajani R, Dmitriev E, Evdokimov V, Frei KM, Gromov A, Goryachev A, Hakonarson H, Hegay T, Khachatryan Z, Khaskhanov R, Kitov E, Kolbina A, Kubatbek T, Kukushkin A, Kukushkin I, Lau N, Margaryan A, Merkyte I, Mertz IV, Mertz VK, Mijiddorj E, Moiyesev V, Mukhtarova G, Nurmukhanbetov B, Orozbekova Z, Panyushkina I, Pieta K, Smrčka V, Shevnina I, Logvin A, Sjogren KG, Štolcova T, Taravella AM, Tashbaeva K, Tkachev A, Tulegenov T, Voyakin D, Yepiskoposyan L, Undrakhbold S, Varfolomeev V, Weber A, Wilson Sayres MA, Kradin N, Allentoft ME, Orlando L, Nielsen R, Sikora M, Heyer E, Kristiansen K, Willerslev E. 137 ancient human genomes from across the Eurasian steppes. Nature, 2018, 563(7729): E16. |
| [26] |
Fan SH, Hansen MEB, Lo Y, Tishkoff SA. Going global by adapting local: a review of recent human adaptation. Science, 2016, 354(6308): 54-59.
pmid: 27846491 |
| [27] | Wang HR, Yang MA, Wangdue S, Lu HL, Chen HH, Li LH, Dong GH, Tsring T, Yuan HB, He W, Ding MY, Wu XH, Li S, Tashi N, Yang T, Yang F, Tong Y, Chen ZJ, He YH, Cao P, Dai QY, Liu F, Feng XT, Wang TY, Yang RW, Ping WJ, Zhang ZX, Gao Y, Zhang M, Wang XJ, Zhang C, Yuan K, Ko AM, Aldenderfer M, Gao X, Xu SH, Fu QM. Human genetic history on the Tibetan Plateau in the past 5100 years. Sci Adv, 2023, 9(11): eadd5582. |
| [28] | Cong PK, Bai WY, Li JC, Yang MY, Khederzadeh S, Gai SR, Li N, Liu YH, Yu SH, Zhao WW, Liu JQ, Sun Y, Zhu XW, Zhao PP, Xia JW, Guan PL, Qian Y, Tao JG, Xu L, Tian G, Wang PY, Xie SY, Qiu MC, Liu KQ, Tang BS, Zheng HF. Genomic analyses of 10,376 individuals in the Westlake BioBank for Chinese (WBBC) pilot project. Nat Commun, 2022, 13(1): 2939. |
| [29] | Chen SQ, Ye CY. Theoretical retrospection on the origin of microblade technology. Acta Anthropologica Sinica, 2019, 38(4): 547-562. |
| 陈胜前, 叶灿阳. 细石叶工艺起源研究的理论反思. 人类学学报, 2019, 38(4): 547-562. | |
| [30] | Ping WQ, Liu YC, Fu QM. Exploring the evolution of archaic humans through sedimentary ancient DNA. Hereditas(Beijing), 2022, 44(5): 362-369. |
| 平婉菁, 刘逸宸, 付巧妹. 沉积物古DNA探秘灭绝古人类演化. 遗传, 2022, 44(5): 362-369. | |
| [31] | Smith O, Dunshea G, Sinding MHS, Fedorov S, Germonpre M, Bocherens H, Gilbert MTP. Ancient RNA from Late Pleistocene permafrost and historical canids shows tissue-specific transcriptome survival. PLoS Biol, 2019, 17(7): e3000166. |
| [32] | Mármol-Sánchez E, Fromm B, Oskolkov N, Pochon Z, Kalogeropoulos P, Eriksson E, Biryukova I, Sekar V, Ersmark E, Andersson B, Dalén L, Friedländer MR. Historical RNA expression profiles from the extinct Tasmanian tiger. Genome Res, 2023, 33(8): 1299-1316. |
| [33] |
Barouch A, Mathov Y, Meshorer E, Yakir B, Carmel L. Reconstructing DNA methylation maps of ancient populations. Nucleic Acids Res, 2024, 52(4): 1602-1612.
doi: 10.1093/nar/gkad1232 pmid: 38261973 |
| [1] | 盛桂莲, 郑铭旻, 肖博, 袁俊霞. 第四纪晚期中国大型哺乳动物古DNA研究进展[J]. 遗传, 2025, 47(1): 46-57. |
| [2] | 郑泽权, 付巧妹, 刘逸宸. 应用古DNA技术探究发酵微生物的适应、演化和驯化历史[J]. 遗传, 2022, 44(5): 414-423. |
| [3] | 平婉菁, 刘逸宸, 付巧妹. 沉积物古DNA探秘灭绝古人类演化[J]. 遗传, 2022, 44(5): 362-369. |
| [4] | 田娇阳, 李玉春, 孔庆鹏, 张亚平. 遗传学视角下东亚人群的起源和演化[J]. 遗传, 2018, 40(10): 814-824. |
| [5] | 王琳淇. 适应性策略:人类致病真菌新生隐球菌的“杀手锏”[J]. 遗传, 2015, 37(5): 436-441. |
| [6] | 盛桂莲,赖旭龙,王,頠. 分子人类学与现代人的起源*[J]. 遗传, 2004, 26(5): 721-728. |
| [7] | 周斌,张思仲,肖翠英,张克兰,张立,李贵星,李萍,刘合焜,郑克勤. 胆固醇7α-羟化酶基因A-204C单核苷酸多态性及其与血浆血脂的关联[J]. 遗传, 2004, 26(3): 0-294. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||

www.chinagene.cn
备案号:京ICP备09063187号-4
总访问:,今日访问:,当前在线:
