Hereditas(Beijing) ›› 2024, Vol. 46 ›› Issue (1): 18-33.doi: 10.16288/j.yczz.23-214
• Review • Previous Articles Next Articles
Genes that escape from X-chromosome inactivation and sexual dimorphism of systemic lupus erythematosus
Qian Ma1,2( ), Shaolan Zhou3(
), Shaolan Zhou3( ), Jie Dang2,4, Zhenghao Huo2,4(
), Jie Dang2,4, Zhenghao Huo2,4( ), Zhanbing Ma2,4(
), Zhanbing Ma2,4( )
)
- 1. School of Life Sciences, Ningxia University, Yinchuan 750021, China
2. Department of Medical Genetics and Cell Biology, College of Basic Medical Sciences, Ningxia Medical University, Yinchuan 750004, China
3. Department of Rheumatology, General Hospital of Ningxia Medical University, Yinchuan 750003, China
4. Key Laboratory of Fertility Preservation and Maintenance (Ningxia Medical University), Ministry of Education, Yinchuan 750004, China
-
Received:2023-10-07Revised:2023-12-11Online:2024-01-20Published:2023-12-13 -
Contact:Zhenghao Huo,Zhanbing Ma E-mail:maqian226@126.com;64758961@qq.com;huozhh@163.com;1784947489@qq.com -
Supported by:National Natural Science Foundation of China(82060301);National Natural Science Foundation of China(81960306);Natural Science Foundation of the Ningxia(22KJB180014);Natural Science Foundation of the Ningxia(2020AAC03116)
Cite this article
Qian Ma, Shaolan Zhou, Jie Dang, Zhenghao Huo, Zhanbing Ma. Genes that escape from X-chromosome inactivation and sexual dimorphism of systemic lupus erythematosus[J]. Hereditas(Beijing), 2024, 46(1): 18-33.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1
SLE sexual dimorphism related XCI escape genes"
| 名称 | 位置 | 表达情况 | 生物学功能 | 参考文献 |
|---|---|---|---|---|
| TLR7 | Xp22.2 | 与健康对照相比,SLE患者表达升高;与男性特纳综合征患者相比,女性患者表达升高 | 调节B细胞增殖、分化和存活,激活自身反应性B细胞,促进自身抗体产生;促进Th1极化;调节CCL5分泌;促进I型干扰素分泌 | [ |
| CD40L | Xq26 | 与男性相比,女性SLE患者B细胞表达升高 | 促进CD4+T细胞反应;诱导幼稚CD8+T细胞活化,促进对效应细胞毒性T淋巴细胞的启动 | [ |
| CXCR3 | Xq13 | 与健康对照相比,SLE患者表达升高;肾脏受累SLE患者表达升高 | 诱导趋化因子受体和Th1免疫反应 | [ |
| IRAK-1 | Xq28 | 与健康对照相比,SLE患者CD4+T细胞表达升高;与男性SLE患者相比,女性患者 T细胞中表达升高 | 激活免疫反应;促进T细胞亚群的分化和激活,进而促进自身免疫反应 | [ |
| CXorf21 | Xp21 | 与男性相比,女性免疫细胞中表达升高 | 特异性激活转录因子IRF5;调节TLR7和TLR9的表达,影响IFN-α、TNF-α、IL-6和趋化因子的表达,影响STAT1和IRF5的磷酸化 | [ |
| [1] |
Zucchi D, Silvagni E, Elefante E, Signorini V, Cardelli C, Trentin F, Schilirò D, Cascarano G, Valevich A, Bortoluzzi A, Tani C. Systemic lupus erythematosus: one year in review 2023. Clin Exp Rheumatol, 2023, 41(5): 997-1008.
doi: 10.55563/clinexprheumatol/4uc7e8 pmid: 37133502 |
| [2] |
Tian JR, Zhang DY, Yao X, Huang YQ, Lu QJ. Global epidemiology of systemic lupus erythematosus: a comprehensive systematic analysis and modelling study. Ann Rheum Dis, 2023, 82(3): 351-356.
doi: 10.1136/ard-2022-223035 |
| [3] |
Ortona E, Pierdominici M, Maselli A, Veroni C, Aloisi F, Shoenfeld Y. Sex-based differences in autoimmune diseases. Ann Ist Super Sanita, 2016, 52(2): 205-212.
doi: 10.4415/ANN_16_02_12 pmid: 27364395 |
| [4] |
Nusbaum JS, Mirza I, Shum J, Freilich RW, Cohen RE, Pillinger MH, Izmirly PM, Buyon JP. Sex differences in systemic lupus erythematosus: epidemiology, clinical considerations, and disease pathogenesis. Mayo Clinic Proceedings, 2020, 95(2): 384-394.
doi: S0025-6196(19)30822-5 pmid: 32029091 |
| [5] |
Christou EAA, Banos A, Kosmara D, Bertsias GK, Boumpas DT. Sexual dimorphism in SLE: above and beyond sex hormones. Lupus, 2019, 28(1): 3-10.
doi: 10.1177/0961203318815768 pmid: 30501463 |
| [6] |
Posynick BJ, Brown CJ. Escape from X-chromosome inactivation: an evolutionary perspective. Front Cell Dev Biol, 2019, 7: 241.
doi: 10.3389/fcell.2019.00241 pmid: 31696116 |
| [7] | Loda A, Collombet S, Heard E. Gene regulation in time and space during X-chromosome inactivation. Nat Rev Mol Cell Biol, 2022, 23(4): 231-249. |
| [8] |
Berletch JB, Yang F, Disteche CM. Escape from X inactivation in mice and humans. Genome Biol, 2010, 11(6): 213.
doi: 10.1186/gb-2010-11-6-213 pmid: 20573260 |
| [9] |
Balaton BP, Brown CJ. Escape artists of the X chromosome. Trends Genet, 2016, 32(6): 348-359.
doi: S0168-9525(16)30002-6 pmid: 27103486 |
| [10] | Lyon M F. Genetic activity of sex chromosomes in somatic cells of mammals. Philos Trans R Soc Lond B Biol Sci, 1970, 259(828): 41-52. |
| [11] |
Schempp W, Meer B. Cytologic evidence for three human X-chromosomal segments escaping inactivation. Hum Genet, 1983, 63(2): 171-174.
pmid: 6682404 |
| [12] |
Disteche CM. Escapees on the X chromosome. Proc Natl Acad Sci USA, 1999, 96(25): 14180-14182.
pmid: 10588671 |
| [13] |
Carrel L, Willard HF. Heterogeneous gene expression from the inactive X chromosome: an X-linked gene that escapes X inactivation in some human cell lines but is inactivated in others. Proc Natl Acad Sci USA, 1999, 96(13): 7364-7369.
doi: 10.1073/pnas.96.13.7364 pmid: 10377420 |
| [14] |
Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature, 2005, 434(7031): 400-404.
doi: 10.1038/nature03479 |
| [15] |
Carrel L, Brown CJ. When the Lyonized (ized chromosome) roars: ongoing expression from an inactive X chromosome. Philos Trans R Soc Lond B Biol Sci, 2017, 372(1733): 20160355.
doi: 10.1098/rstb.2016.0355 |
| [16] |
Qi SH, Al Mamun A, Ngwa C, Romana S, Ritzel R, Arnold AP, McCullough LD, Liu FD. X chromosome escapee genes are involved in ischemic sexual dimorphism through epigenetic modification of inflammatory signals. Neuroinflammation, 2021, 18(1): 70.
doi: 10.1186/s12974-021-02120-3 |
| [17] |
Al Nadaf S, Deakin JE, Gilbert C, Robinson TJ, Graves JAM, Waters PD. A cross-species comparison of escape from X inactivation in Eutheria: implications for evolution of X chromosome inactivation. Chromosome, 2012, 121(1): 71-78.
doi: 10.1007/s00412-011-0343-8 |
| [18] |
Yang F, Babak T, Shendure J, Disteche CM. Global survey of escape from X inactivation by RNA- sequencing in mouse. Genome Res, 2010, 20(5): 614-622.
doi: 10.1101/gr.103200.109 pmid: 20363980 |
| [19] |
Wang X, Douglas KC, Vandeberg JL, Clark AG, Samollow PB. Chromosome-wide profiling of X-chromosome inactivation and epigenetic states in fetal brain and placenta of the opossum, Monodelphis domestica. Genome Res, 2014, 24(1): 70-83.
doi: 10.1101/gr.161919.113 pmid: 24065774 |
| [20] |
Whitworth D J, Pask AJ. The X factor: X chromosome dosage compensation in the evolutionarily divergent monotremes and marsupials. Semin Cell Dev Biol, 2016, 56: 117-121.
doi: S1084-9521(16)30006-4 pmid: 26806635 |
| [21] | Balaton BP, Cotton AM, Brown CJ. Derivation of consensus inactivation status for X-linked genes from genome-wide studies. Biol Sex Diff, 2015, 6: 35. |
| [22] |
Zhang YC, Castillo-Morales A, Jiang M, Zhu YF, Hu LD, Urrutia AO, Kong XY, Hurst LD. Genes that escape X-inactivation in humans have high intraspecific variability in expression,are associated with mental impairment but are not slow evolving. Mol Biol Evol, 2013, 30(12): 2588-2601.
doi: 10.1093/molbev/mst148 |
| [23] |
Tukiainen T, Villani AC, Yen A, Rivas MA, Marshall JL, Satija R, Aguirre M, Gauthier L, Fleharty M, Kirby A, Cummings BB, Castel SE, Karczewski KJ, Aguet F, Byrnes A, GTEx Consortium, Laboratory, Data Analysis & Coordinating Center (LDACC)—Analysis Working Group, Statistical Methods groups—Analysis Working Group, Enhancing GTEx (eGTEx) groups, NIH Common Fund, NIH/NCI, NIH/NHGRI, NIH/NIMH, NIH/NIDA, Biospecimen Collection Source Site—NDRI, Biospecimen Collection Source Site—RPCI, Biospecimen Core Resource—VARI, Brain Bank Repository—University of Miami Brain Endowment Bank, Leidos Biomedical— Project Management, ELSI Study, Genome Browser Data Integration &Visualization—EBI, Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz, Lappalainen T, Regev A, Ardlie KG, Hacohen N, MacArthur DG. Landscape of X chromosome inactivation across human tissues. Nature, 2017, 550: 244-248.
doi: 10.1038/nature24265 |
| [24] |
Garieri M, Stamoulis G, Blanc X, Falconnet E, Ribaux P, Borel C, Santoni F, Antonarakis SE. Extensive cellular heterogeneity of X inactivation revealed by single-cell allele-specific expression in human fibroblasts. Proc Natl Acad Sci USA, 2018, 115(51): 13015-13020.
doi: 10.1073/pnas.1806811115 pmid: 30510006 |
| [25] |
Zito A, Roberts AL, Visconti A, Rossi N, Andres-Ejarque R, Nardone S, El-Sayed Moustafa JS, Falchi M, Small KS. Escape from X-inactivation in twins exhibits intra- and inter-individual variability across tissues and is heritable. PLoS Genet, 2023, 19(2): e1010556.
doi: 10.1371/journal.pgen.1010556 |
| [26] |
Libert C, Dejager L, Pinheiro I. The X chromosome in immune functions: when a chromosome makes the difference. Nat Rev Immunol, 2010, 10(8): 594-604.
doi: 10.1038/nri2815 pmid: 20651746 |
| [27] |
Neri G, Schwartz CE, Lubs HA, Stevenson RE. X-linked intellectual disability update 2017. Am J Med Genet A, 2018, 176(6): 1375-1388.
doi: 10.1002/ajmg.a.38710 pmid: 29696803 |
| [28] |
Dunford A, Weinstock DM, Savova V, Schumacher SE, Cleary JP, Yoda A, Sullivan TJ, Hess JM, Gimelbrant AA, Beroukhim R, Lawrence MS, Getz G, Lane AA. Tumor-suppressor genes that escape from X-inactivation contribute to cancer sex bias. Nat Genet, 2017, 49(1): 10-16.
doi: 10.1038/ng.3726 pmid: 27869828 |
| [29] |
Clement-Jones M, Schiller S, Rao E, Blaschke RJ, Zuniga A, Zeller R, Robson SC, Binder G, Glass I, Strachan T, Lindsay S, Rappold GA. The short stature homeobox gene SHOX is involved in skeletal abnormalities in Turner syndrome. Hum Mol Genet, 2000, 9(5): 695-702.
doi: 10.1093/hmg/9.5.695 pmid: 10749976 |
| [30] |
Sauteraud R, Stahl JM, James J, Englebright M, Chen F, Zhan XW, Carrel L, Liu DJ. Inferring genes that escape X-Chromosome inactivation reveals important contribution of variable escape genes to sex-biased diseases. Genome Res, 2021, 31(9): 1629-1637.
doi: 10.1101/gr.275677.121 pmid: 34426515 |
| [31] | Augui S, Nora EP, Heard E. Regulation of X-chromosome inactivation by the X-inactivation centre. Nat Rev Genet, 2011, 12(6): 429-442. |
| [32] |
Pandya-Jones A, Markaki Y, Serizay J, Chitiashvili T, Mancia Leon WR, Damianov A, Chronis C, Papp B, Chen CK, McKee R, Wang XJ, Chau A, Sabri S, Leonhardt H, Zheng SK, Guttman M, Black DL, Plath K. A protein assembly mediates Xist localization and gene silencing. Nature, 2020, 587(7832): 145-151.
doi: 10.1038/s41586-020-2703-0 |
| [33] |
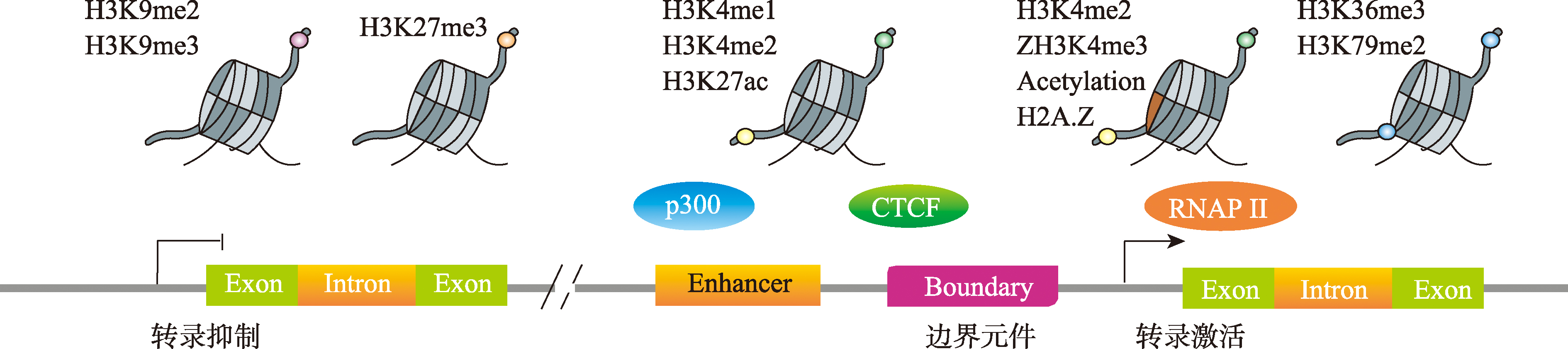
Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet, 2011, 12(1): 7-18.
doi: 10.1038/nrg2905 pmid: 21116306 |
| [34] |
Loda A, Heard E. Xist RNA in action: past, present, and future. PLoS Genet, 2019, 15: e1008333.
doi: 10.1371/journal.pgen.1008333 |
| [35] |
Wang ZZ, Cai HR, Zhao EH, Cui HJ. The diverse roles of histone demethylase KDM4B in normal and cancer development and progression. Front Cell Dev Biol, 2022, 9: 790129.
doi: 10.3389/fcell.2021.790129 |
| [36] |
Sadreyev RI, Yildirim E, Pinter SF, Lee JT. Bimodal quantitative relationships between histone modifications for X-linked and autosomal loci. Proc Natl Acad Sci USA, 2013, 110(17): 6949-6954.
doi: 10.1073/pnas.1216449110 pmid: 23564346 |
| [37] |
Kelsey AD, Yang C, Leung D, Minks J, Dixon- McDougall T, Baldry SEL, Bogutz AB, Lefebvre L, Brown CJ. Impact of flanking chromosomal sequences on localization and silencing by the human non-coding RNA XIST. Genom Biol, 2015, 16: 208.
doi: 10.1186/s13059-015-0774-2 |
| [38] |
Brockdorff N. Polycomb complexes in X chromosome inactivation. Philos Trans R Soc Lond B Biol Sci, 2017, 372(1733): 20170021.
doi: 10.1098/rstb.2017.0021 |
| [39] |
Cerase A, Smeets D, Tang YA, Gdula M, Kraus F, Spivakov M, Moindrot B, Leleu M, Tattermusch A, Demmerle J, Nesterova TB, Green C, Otte AP, Schermelleh L, Brockdorff N. Spatial separation of Xist RNA and polycomb proteins revealed by superresolution microscopy. Proc Natl Acad Sci USA, 2014, 111(6): 2235-2240.
doi: 10.1073/pnas.1312951111 pmid: 24469834 |
| [40] |
Simon MD, Pinter SF, Fang R, Sarma K, Rutenberg- Schoenberg M, Bowman SK, Kesner BA, Maier VK, Kingston RE, Lee JT. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature, 2013, 504(7480): 465-469.
doi: 10.1038/nature12719 |
| [41] |
Brinkman AB, Roelofsen T, Pennings SW, Martens JHA, Jenuwein T, Stunnenberg HG. Histone modification patterns associated with the human X chromosome. EMBO Rep, 2006, 7(6): 628-634.
doi: 10.1038/sj.embor.7400686 pmid: 16648823 |
| [42] |
Tang SJ. Chromatin organization by repetitive elements (CORE): a genomic principle for the higher-order structure of chromosomes. Genes, 2011, 2: 502-515.
doi: 10.3390/genes2030502 |
| [43] |
Wang Z, Willard HF, Mukherjee S, Furey TS. Evidence of influence of genomic DNA sequence on human X chromosome inactivation. PLoS Comput Biol, 2006, 2(9): e113.
doi: 10.1371/journal.pcbi.0020113 pmid: 16948528 |
| [44] |
Lyon MF. X-chromosome inactivation: a repeat hypothesis. Cytogenet Cell Genet, 1998, 80(1-4): 133-137.
doi: 10.1159/000014969 pmid: 9678347 |
| [45] | Ross MT, Grafham DV, Coffey AJ, Scherer S, McLay K, Muzny D, Platzer M, Howell GR, Burrows C, Bird CP, Frankish A, Lovell FL, Howe KL, Ashurst JL, Fulton RS, Sudbrak R, Wen GP, Jones MC, Hurles ME, Andrews TD, Scott CE, Searle S, Ramser J, Whittaker A, Deadman R, Carter NP, Hunt SE, Chen R, Cree A, Gunaratne P, Havlak P, Hodgson A, Metzker ML, Richards S, Scott G, Steffen D, Sodergren E, Wheeler DA, Worley KC, Ainscough R, Ambrose KD, Ansari-Lari MA, Aradhya S, Ashwell RIS, Babbage AK, Bagguley CL, Ballabio A, Banerjee R, Barker GE, Barlow KF, Barrett IP, Bates KN, Beare DM, Beasley H, Beasley O, Beck A, Bethel G, Blechschmidt K, Brady N, Bray-Allen S, Bridgeman AM, Brown AJ, Brown MJ, Bonnin D, Bruford EA, Buhay C, Burch P, Burford D, Burgess J, Burrill W, Burton J, Bye JM, Carder C, Carrel L, Chako J, Chapman JC, Chavez D, Chen E, Chen G, Chen Y, Chen ZJ, Chinault C, Ciccodicola A, Clark SY, Clarke G, Clee CM, Clegg S, Clerc-Blankenburg K, Clifford K, Cobley V, Cole CG, Conquer JS, Corby N, Connor RE, David R, Davies J, Davis C, Davis J, Delgado O, Deshazo D, Dhami P, Ding Y, Dinh H, Dodsworth S, Draper H, Dugan-Rocha S, Dunham A, Dunn M, Durbin KJ, Dutta I, Eades T, Ellwood M, Emery-Cohen A, Errington H, Evans KL, Faulkner L, Francis F, Frankland J, Fraser AE, Galgoczy P, Gilbert J, Gill R, Glöckner G, Gregory SG, Gribble S, Griffiths C, Grocock R, Gu Y, Gwilliam R, Hamilton C, Hart EA, Hawes A, Heath PD, Heitmann K, Hennig S, Hernandez J, Hinzmann B, Ho S, Hoffs M, Howden PJ, Huckle EJ, Hume J, Hunt PJ, Hunt AR, Isherwood J, Jacob L, Johnson D, Jones S, de Jong PJ, Joseph SS, Keenan S, Kelly S, Kershaw JK, Khan Z, Kioschis P, Klages S, Knights AJ, Kosiura A, Kovar-Smith C, Laird GK, Langford C, Lawlor S, Leversha M, Lewis L, Liu W, Lloyd C, Lloyd DM, Loulseged H, Loveland JE, Lovell JD, Lozado R, Lu J, Lyne R, Ma J, Maheshwari M, Matthews LH, McDowall J, McLaren S, McMurray A, Meidl P, Meitinger T, Milne S, Miner G, Mistry SL, Morgan M, Morris S, Müller I, Mullikin JC, Nguyen N, Nordsiek G, Nyakatura G, O'Dell CN, Okwuonu G, Palmer S, Pandian R, Parker D, Parrish J, Pasternak S, Patel D, Pearce AV, Pearson DM, Pelan SE, Perez L, Porter KM, Ramsey Y, Reichwald K, Rhodes S, Ridler KA, Schlessinger D, Schueler MG, Sehra HK, Shaw-Smith C, Shen H, Sheridan EM, Shownkeen R, Skuce CD, Smith ML, Sotheran EC, Steingruber HE, Steward CA, Storey R, Swann RM, Swarbreck D, Tabor PE, Taudien S, Taylor T, Teague B, Thomas K, Thorpe A, Timms K, Tracey A, Trevanion S, Tromans AC, d'Urso M, Verduzco D, Villasana D, Waldron L, Wall M, Wang Q, Warren J, Warry GL, Wei X, West A, Whitehead SL, Whiteley MN, Wilkinson JE, Willey DL, Williams G, Williams L, Williamson A, Williamson H, Wilming L, Woodmansey RL, Wray PW, Yen J, Zhang J, Zhou J, Zoghbi H, Zorilla S, Buck D, Reinhardt R, Poustka A, Rosenthal A, Lehrach H, Meindl A, Minx PJ, Hillier LW, Willard HF, Wilson RK, Waterston RH, Rice CM, Vaudin M, Coulson A, Nelson DL, Weinstock G, Sulston JE, Durbin R, Hubbard T, Gibbs RA, Beck S, Rogers J, Bentley DR. The DNA sequence of the human X chromosome. Nature, 2005, 434: 325-337. |
| [46] |
Chow JC, Ciaudo C, Fazzari MJ, Mise N, Servant N, Glass JL, Attreed M, Avner P, Wutz A, Barillot E, Greally JM, Voinnet O, Heard E. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell, 2016, 166(3): 782.
doi: S0092-8674(16)30920-5 pmid: 27471971 |
| [47] |
Li SY, Shen XH. Long interspersed nuclear element 1 and B1/Alu repeats blueprint genome compartmentalization. Curr Opin Genet Dev, 2023, 80: 102049.
doi: 10.1016/j.gde.2023.102049 |
| [48] |
Nguyen DK, Yang F, Kaul R, Alkan C, Antonellis A, Friery KF, Zhu BL, de Jong PJ, Disteche CM. Clcn4-2 genomic structure differs between the X locus in Mus spretus and the autosomal locus in Mus musculus: AT motif enrichment on the X. Genome Res, 2011, 21(3): 402-409.
doi: 10.1101/gr.108563.110 pmid: 21282478 |
| [49] |
Helbig R, Fackelmayer FO. Scaffold attachment factor A (SAF-A) is concentrated in inactive X chromosome territories through its RGG domain. Chromosoma, 2003, 112(4): 173-182.
pmid: 14608463 |
| [50] |
Agrelo R, Souabni A, Novatchkova M, Haslinger C, Leeb M, Komnenovic V, Kishimoto H, Gresh L, Kohwi- Shigematsu T, Kenner L, Wutz A. SATB1 defines the developmental context for gene silencing by Xist in lymphoma and embryonic cells. Dev Cell, 2009, 16(4): 507-516.
doi: 10.1016/j.devcel.2009.03.006 pmid: 19386260 |
| [51] |
Kohwi-Shigematsu T, Kohwi Y, Takahashi K, Richards HW, Ayers SD, Han HJ, Cai ST. SATB1-mediated functional packaging of chromatin into loops. Methods, 2012, 58(3): 243-254.
doi: 10.1016/j.ymeth.2012.06.019 pmid: 22782115 |
| [52] |
Galande S, Purbey PK, Notani D, Kumar PP.The third dimension of gene regulation: organization of dynamic chromatin loopscape by SATB1. Curr Opin Genet Dev, 2007, 17(5): 408-414.
doi: 10.1016/j.gde.2007.08.003 pmid: 17913490 |
| [53] |
Hasegawa Y, Brockdorff N, Kawano S, Tsutui K, Tsutui K, Nakagawa S. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev Cell, 2010, 19(3): 469-476.
doi: 10.1016/j.devcel.2010.08.006 pmid: 20833368 |
| [54] | Sanborn AL, Rao SS, Huang SC, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J, Geeting KP, Gnirke A, Melnikov A, McKenna D, Stamenova EK, Lander ES, Aiden EL. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci USA, 2015, 112(47): E6456-E6465. |
| [55] |
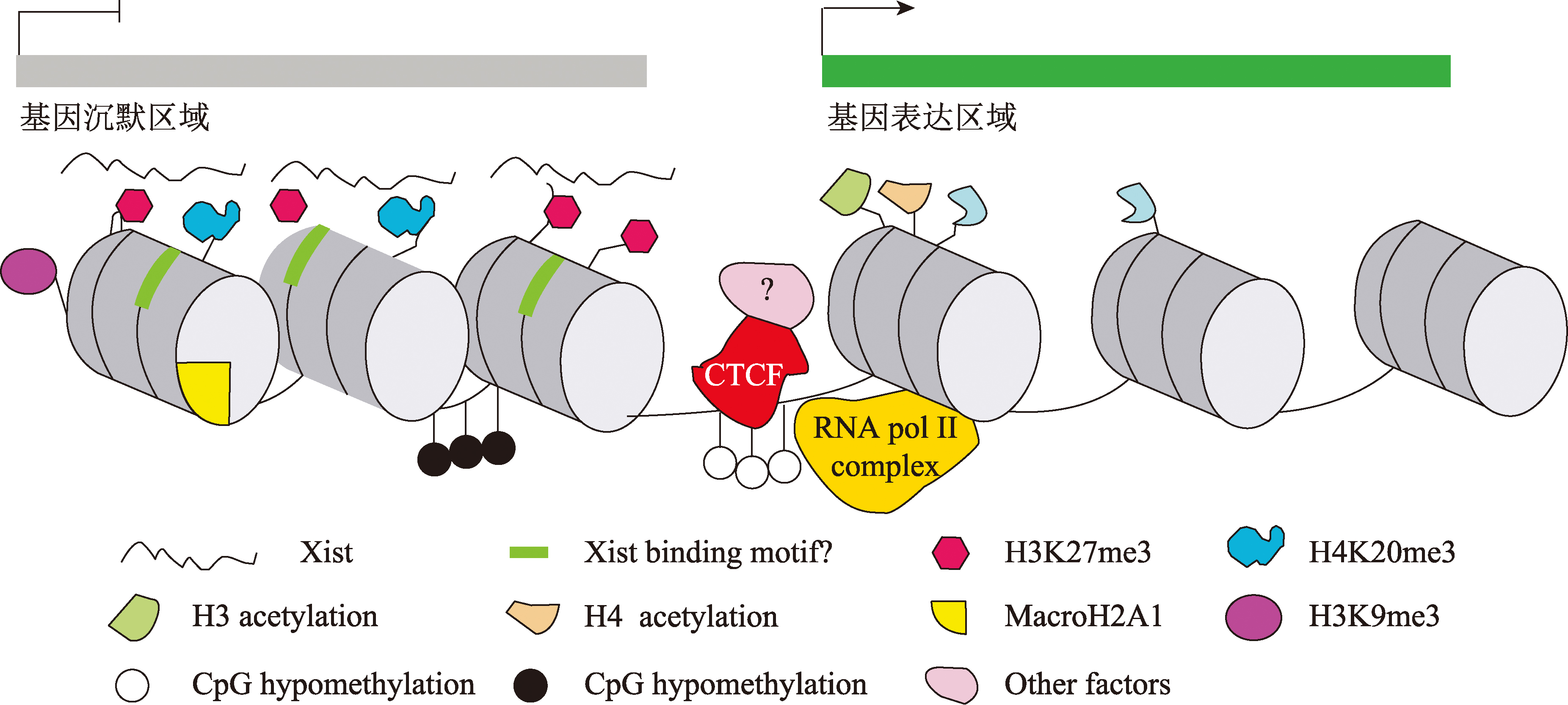
Filippova GN, Cheng MK, Moore JM, Truong JP, Hu YJ, Nguyen DK, Tsuchiya KD, Disteche CM. Boundaries between chromosomal domains of X inactivation and escape bind CTCF and lack CpG methylation during early development. Dev Cell, 2005, 8(1): 31-42.
doi: 10.1016/j.devcel.2004.10.018 pmid: 15669143 |
| [56] | Fang H, Tronco AR, Bonora G, Nguyen T, Thakur J, Berletch JB, Filippova GN, Henikoff S, Shendure J, Noble WS, Disteche CM, Deng XX. CTCF-mediated insulation and chromatin environment modulate Car5b escape from X inactivation. bioRXiv, 2023. |
| [57] |
Chen CY, Shi WQ, Balaton BP, Matthews AM, Li YF, Arenillas DJ, Mathelier A, Itoh M, Kawaji H, Lassmann T, Hayashizaki Y, Carninci P, Forrest ARR, Brown CJ, Wasserman WW. YY1 binding association with sex- biased transcription revealed through X-linked transcript levels and allelic binding analyses. Sci Rep, 2016, 6: 37324.
doi: 10.1038/srep37324 |
| [58] |
Li L, Williams P, Ren WD, Wang MY, Gao Z, Miao WL, Huang M, Song JK, Wang YS. YY1 interacts with guanine quadruplexes to regulate DNA looping and gene expression. Nat Chem Biol, 2021, 17(2): 161-168.
doi: 10.1038/s41589-020-00695-1 pmid: 33199912 |
| [59] |
Sun S, Del Rosario BC, Szanto A, Ogawa Y, Jeon Y, Lee JT. Jpx RNA activates Xist by evicting CTCF. Cell, 2013, 153(7): 1537-1551.
doi: 10.1016/j.cell.2013.05.028 pmid: 23791181 |
| [60] |
Oh HJ, Aguilar R, Kesner B, Lee HG, Kriz AJ, Chu HP, Lee JT. Jpx RNA regulates CTCF anchor site selection and formation of chromosome loops. Cell, 2021, 184(25): 6157-6173.e24.
doi: 10.1016/j.cell.2021.11.012 pmid: 34856126 |
| [61] | Jiwrajka N, Anguera MC. The X in sex-biased immunity and autoimmune rheumatic disease. J Exp Med, 2022, 219(6): e20211487. |
| [62] |
Miquel CH, Faz-Lopez B, Guéry JC. Influence of X chromosome in sex-biased autoimmune diseases. J Autoimmun, 2023, 137: 102992.
doi: 10.1016/j.jaut.2023.102992 |
| [63] |
Berletch JB, Yang F, Xu J, Carrel L, Disteche CM. Genes that escape from X inactivation. Hum Genet, 2011, 130(2): 237-245.
doi: 10.1007/s00439-011-1011-z pmid: 21614513 |
| [64] |
Sarmiento L, Svensson J, Barchetta I, Mousavi MJ, Mahmoudi M, Ghotloo S. Escape from X chromosome inactivation and female bias of autoimmune diseases. Mol Med, 2020, 26(1): 127.
doi: 10.1186/s10020-020-00256-1 pmid: 33297945 |
| [65] |
Syrett CM, Anguera MC. When the balance is broken: X linked gene dosage from two X chromosomes and female-biased autoimmunity. J Leukoc Biol, 2019, 106(4): 919-932.
doi: 10.1002/JLB.6RI0319-094R |
| [66] |
Sarmiento L, Svensson J, Barchetta I, Giwercman A, Cilio CM. Copy number of the X-linked genes TLR7 and CD40L influences innate and adaptive immune responses. Scand J Immunol, 2019, 90(2): e12776.
doi: 10.1111/sji.2019.90.issue-2 |
| [67] | Souyris M, Cenac C, Azar P, Daviaud D, Canivet A, Grunenwald S, Pienkowski C, Chaumeil J, Mejía J, Guéry JC. TLR7 escapes X chromosome inactivation in immune cells. Sci Immunol, 2018, 3(19): eaap8855. |
| [68] |
Fillatreau S, Manfroi B, Dörner T. Toll-like receptor signalling in B cells during systemic lupus erythematosus. Nat Rev Rheumatol, 2021, 17(2): 98-108.
doi: 10.1038/s41584-020-00544-4 pmid: 33339987 |
| [69] |
Souyris M, Mejía JE, Chaumeil J, Guéry JC. Female predisposition to TLR7-driven autoimmunity: gene dosage and the escape from X chromosome inactivation. Semin Immunopathol, 2019, 41(2): 153-164.
doi: 10.1007/s00281-018-0712-y pmid: 30276444 |
| [70] |
Murphy ED, Roths JB. A Y chromosome associated factor in strain BXSB producing accelerated autoimmunity and lymphoproliferation. Arthritis Rheum, 1979, 22(11): 1188-1194.
doi: 10.1002/art.v22:11 |
| [71] |
Merino R, Fossati L, Izui S. The lupus-prone BXSB strain: the Yaa gene model of systemic lupus erythematosus. Springer Semin Immunopathol, 1992, 14(2): 141-157.
pmid: 1475741 |
| [72] |
Izui S, Iwamoto M, Fossati L, Merino R, Takahashi S, Ibnou-Zekri N. The Yaa gene model of systemic lupus erythematosus. Immunol Rev, 1995, 144: 137-156.
pmid: 7590811 |
| [73] |
Amano H, Amano E, Moll T, Marinkovic D, Ibnou-Zekri N, Martinez-Soría E, Semac I, Wirth T, Nitschke L, Izui S. The Yaa mutation promoting murine lupus causes defective development of marginal zone B cells. J Immunol, 2003, 170(5): 2293-2301.
doi: 10.4049/jimmunol.170.5.2293 pmid: 12594250 |
| [74] |
Pisitkun P, Deane JA, Difilippantonio MJ, Tarasenko T, Satterthwaite AB, Bolland S. Autoreactive B cell responses to RNA-related antigens due to TLR7 gene duplication. Science, 2006, 312(5780): 1669-1672.
doi: 10.1126/science.1124978 pmid: 16709748 |
| [75] |
Slae M, Heshin-Bekenstein M, Simckes A, Heimer G, Engelhard D, Eisenstein EM. Female polysomy-X and systemic lupus erythematosus. Semin Arthritis Rheum, 2014, 43(4): 508-512.
doi: 10.1016/j.semarthrit.2013.07.014 |
| [76] |
Harris VM, Sharma R, Cavett J, Kurien BT, Liu K, Koelsch KA, Rasmussen A, Radfar L, Lewis D, Stone DU, Kaufman CE, Li SB, Segal B, Wallace DJ, Weisman MH, Venuturupalli S, Kelly JA, Alarcon-Riquelme ME, Pons-Estel B, Jonsson R, Lu XL, Gottenberg JE, Anaya JM, Cunninghame-Graham DS, Huang AJW, Brennan MT, Hughes P, Alevizos I, Miceli-Richard C, Keystone EC, Bykerk VP, Hirschfield G, Xie G, Siminovitch KA, Ng WF, Nordmark G, Bucher SM, Eriksson P, Omdal R, Rhodus NL, Rischmueller M, Rohrer M, Wahren- Herlenius M, Witte T, Mariette X, Lessard CJ, Harley JB, Sivils KL, Scofield RH. Klinefelter’s syndrome (47,XXY) is in excess among men with Sjögren’s syndrome. Clin Immunol, 2016, 168: 25-29.
doi: 10.1016/j.clim.2016.04.002 |
| [77] |
Sawalha AH, Harley JB, Scofield RH. Autoimmunity and Klinefelter’s syndrome: when men have two X chromosomes. J Autoimmun, 2009, 33(1): 31-34.
doi: 10.1016/j.jaut.2009.03.006 pmid: 19464849 |
| [78] |
Invernizzi P, Miozzo M, Oertelt-Prigione S, Meroni PL, Persani L, Selmi C, Battezzati PM, Zuin M, Lucchi S, Marasini B, Zeni S, Watnik M, Tabano S, Maitz S, Pasini S, Gershwin ME, Podda M. X monosomy in female systemic lupus erythematosus. Ann N Y Acad Sci, 2007, 1110: 84-91.
doi: 10.1196/annals.1423.010 |
| [79] | Liu K, Kurien BT, Zimmerman SL, Kaufman KM, Taft DH, Kottyan LC, Lazaro S, Weaver CA, Ice JA, Adler AJ, Chodosh J, Radfar L, Rasmussen A, Stone DU, Lewis DM, Li SB, Koelsch KA, Igoe A, Talsania M, Kumar J, Maier-Moore JS, Harris VM, Gopalakrishnan R, Jonsson R, Lessard JA, Lu XL, Gottenberg JE, Anaya JM, Cunninghame-Graham DS, Huang AJW, Brennan MT, Hughes P, Illei GG, Miceli-Richard C, Keystone EC, Bykerk VP, Hirschfield G, Xie G, Ng WF, Nordmark G, Eriksson P, Omdal R, Rhodus NL, Rischmueller M, Rohrer M, Segal BM, Vyse TJ, Wahren-Herlenius M, Witte T, Pons-Estel B, Alarcon-Riquelme ME, Guthridge JM, James JA, Lessard CJ, Kelly JA, Thompson SD, Gaffney PM, Montgomery CG, Edberg JC, Kimberly RP, Alarcón GS, Langefeld CL, Gilkeson GS, Kamen DL, Tsao BP, McCune WJ, Salmon JE, Merrill JT, Weisman MH, Wallace DJ, Utset TO, Bottinger EP, Amos CI, Siminovitch KA, Mariette X, Sivils KL, Harley JB, Scofield RH. X Chromosome dose and sex bias in autoimmune diseases: increased prevalence of 47,XXX in systemic lupus erythematosus and Sjögren’s syndrome. Arthritis Rheumatol, 2016, 68(5): 1290-1300. |
| [80] |
Koshy M, Berger D, Crow MK. Increased expression of CD40 ligand on systemic lupus erythematosus lymphocytes. J Clin Invest, 1996, 98: 826-837.
doi: 10.1172/JCI118855 pmid: 8698875 |
| [81] |
Desai-Mehta A, Lu L, Ramsey-Goldman R, Datta SK. Hyperexpression of CD40 ligand by B and T cells in human lupus and its role in pathogenic autoantibody production. J Clin Invest, 1996, 97(7): 2063-2073.
doi: 10.1172/JCI118643 |
| [82] |
Hewagama A, Gorelik G, Patel D, Liyanarachchi P, Liyanarachchi P, McCune WJ, Somers E, Gonzalez- Rivera T, Michigan Lupus Cohort, Strickland F, Richardson B. Overexpression of X-linked genes in T cells from women with lupus. J Autoimmun, 2013, 41: 60-71.
doi: 10.1016/j.jaut.2012.12.006 pmid: 23434382 |
| [83] |
Ramanujam M, Steffgen J, Visvanathan S, Mohan C, Fine JS, Putterman C. Phoenix from the flames: rediscovering the role of the CD40-CD40L pathway in systemic lupus erythematosus and lupus nephritis. Autoimmun Rev, 2020, 19(11): 102668.
doi: 10.1016/j.autrev.2020.102668 |
| [84] |
Vordenbäumen S, Sokolowski A, Rosenbaum A, Gebhard C, Raithel J, Düsing C, Chehab G, Richter JG, Brinks R, Rehli M, Schneider M. Methyl donor micronutrients, CD40-ligand methylation and disease activity in systemic lupus erythematosus: a cross-sectional association study. Lupus, 2021, 30(11): 1773-1780.
doi: 10.1177/09612033211034559 |
| [85] |
Lu QJ, Wu AL, Tesmer L, Ray D, Yousif N, Richardson B. Demethylation of CD40LG on the inactive X in T cells from women with lupus. J Immunol, 2007, 179(9): 6352-6358.
pmid: 17947713 |
| [86] |
Lacotte S, Brun S, Muller S, Dumortier H. CXCR3, inflammation, and autoimmune diseases. Ann N Y Acad Sci, 2009, 1173: 310-317.
doi: 10.1111/nyas.2009.1173.issue-1 |
| [87] |
Oghumu S, Varikuti S, Stock JC, Volpedo G, Saljoughian N, Terrazas CA, Satoskar AR. Cutting Edge: CXCR3 escapes X chromosome inactivation in T cells during infection: potential implications for sex differences in immune responses. J Immunol, 2019, 203(4): 789-794.
doi: 10.4049/jimmunol.1800931 pmid: 31253729 |
| [88] | Barbi J, Oghumu S, Lezama-Davila CM, Satoskar AR. IFN-gamma and STAT1 are required for efficient induction of CXC chemokine receptor 3 (CXCR3) on CD4+ but not CD8+ T cells. Blood, 2007, 110: 2215-2216. |
| [89] |
Wang GJ, Sun Y, Jiang YS, Li SZ, Liu YH, Yuan YY, Nie H. CXCR3 deficiency decreases autoantibody production by inhibiting aberrant activated T follicular helper cells and B cells in lupus mice. Mol Immunol, 2023, 156: 39-47.
doi: 10.1016/j.molimm.2023.02.009 pmid: 36889185 |
| [90] |
Menke J, Zeller GC, Kikawada E, Means TK, Huang XR, Lan HY, Lu B, Farber J, Luster AD, Kelley VR. CXCL9, but not CXCL10, promotes CXCR3-dependent immune- mediated kidney disease. J Am Soc Nephrol, 2008, 19: 1177-1189.
doi: 10.1681/ASN.2007111179 |
| [91] |
Steinmetz OM, Turner JE, Paust HJ, Lindner M, Peters A, Heiss K, Velden J, Hopfer H, Fehr S, Krieger T, Mey er-Schwesinger C, Meyer TN, Helmchen U, Mittrücker HW, Stahl RAK, Panzer U. CXCR3 mediates renal Th1 and Th17 immune response in murine lupus nephritis. J Immunol, 2009, 183: 4693-4704.
doi: 10.4049/jimmunol.0802626 pmid: 19734217 |
| [92] | Fan HY, Dong GJ, Zhao GF, Liu F, Yao GH, Zhu YC, Hou YJ. Gender differences of B cell signature in healthy subjects underlie disparities in incidence and course of SLE related to estrogen. Immunol Res, 2014, 2014: 814598. |
| [93] | Im CH, Park JA, Kim JY, Lee EY, Lee EB, Kim Y, Song YW. CXCR3 polymorphism is associated with male gender and pleuritis in patients with systemic lupus erythematosus. Hum Immunol, 2014, 75(5): 466-469. |
| [94] |
Enghard P, Humrich JY, Rudolph B, Rosenberger S, Biesen R, Kuhn A, Manz R, Hiepe F, Radbruch A, Burmester GR, Riemekasten G. CXCR3+CD4+T cells are enriched in inflamed kidneys and urine and provide a new biomarker for acute nephritis flares in systemic lupus erythematosus patients. Arthritis Rheum, 2009, 60: 199-206.
doi: 10.1002/art.v60:1 |
| [95] |
Cooke EL, Uings IJ, Xia CL, Woo P, Ray KP. Functional analysis of the interleukin-1-receptor-associated kinase (IRAK-1) in interleukin-1 beta-stimulated nuclear factor kappa B (NF-kappa B) pathway activation: IRAK-1 associates with the NF-kappa B essential modulator (NEMO) upon receptor stimulation. Biochem J, 2001, 359: 403-410.
doi: 10.1042/0264-6021:3590403 pmid: 11583588 |
| [96] |
Li-Weber M, Giaisi M, Baumann S, Pálf K, Krammer PH. NF-kappa B synergizes with NF-AT and NF-IL6 in activation of the IL-4 gene in T cells. Eur J Immunol, 2004, 34: 1111-1118.
pmid: 15048722 |
| [97] |
Ruan QG, Kameswaran V, Zhang Y, Zheng SJ, Sun J, Wang JM, DeVirgiliis J, Liou HC, Beg AA, Chen YH. The Th17 immune response is controlled by the Rel-RORγ-RORγ T transcriptional aXis. J Exp Med, 2011, 208: 2321-2333.
doi: 10.1084/jem.20110462 |
| [98] |
Oh H, Ghosh S. NF-kappa B: roles and regulation in different CD4(+) T-cell subsets. Immunol Rev, 2013, 252: 41-51.
doi: 10.1111/imr.2013.252.issue-1 |
| [99] |
Bronson PG, Chaivorapol C, Ortmann W, Behrens TW, Graham RR. The genetics of type I interferon in systemic lupus erythematosus. Curr Opin Immunol, 2012, 24(5): 530-537.
doi: 10.1016/j.coi.2012.07.008 pmid: 22889593 |
| [100] |
Jacob CO, Zhu JK, Armstrong DL, Yan M, Han J, Zhou XJ, Thomas JA, Reiff A, Myones BL, Ojwang JO, Kaufman KM, Klein-Gitelman M, McCurdy D, Wagner-Weiner L, Silverman E, Ziegler J, Kelly JA, Merrill JT, Harley JB, Ramsey-Goldman R, Vila LM, Bae SC, Vyse TJ, Gilkeson GS, Gaffney PM, Moser KL, Langefeld CD, Zidovetzki R, Mohan C. Identifciation of IRAK1 as a risk gene with critical role in the pathogenesis of systemic lupus erythematosus. Proc Natl Acad Sci USA, 2009, 106: 6256-6261.
doi: 10.1073/pnas.0901181106 |
| [101] |
Li MF, Yu DT, Ni B, Hao F. Interleukin-1 receptor associated kinase 1 is a potential therapeutic target of anti-inflammatory therapy for systemic lupus erythematosus. Mol Immunol, 2017, 87: 94-101.
doi: S0161-5890(17)30089-5 pmid: 28431280 |
| [102] |
Li MF, Yu DT, Wang Y, Luo N, Han GM, Yang B. Interferon-α activates interleukin-1 receptor-associated kinase 1 to induce regulatory T-cell apoptosis in patients with systemic lupus erythematosus. J Dermatol, 2021, 48(8): 1172-1185.
doi: 10.1111/jde.v48.8 |
| [103] |
Zhou Z, Tian ZQ, Zhang MJ, Zhang YX, Ni B, Hao F. Upregulated IL-1 receptor-associated kinase 1 (IRAK1) in systemic lupus erythematosus: IRAK1 inhibition represses Th17 differentiation with therapeutic potential. Immunol Invest, 2018, 47(5): 468-483.
doi: 10.1080/08820139.2018.1458105 pmid: 29611775 |
| [104] |
David A, Trigunaite A, Macleod MK, Johnson AC, Marrack P, Jørgensen TN. Intrinsic autoimmune capacities of hematopoietic cells from female New Zealand hybrid mice. Genes Immun, 2014, 15(3): 153-161.
doi: 10.1038/gene.2014.2 pmid: 24477163 |
| [105] |
Odhams CA, Roberts AL, Vester SK, Duarte CST, Beales CT, Clarke AJ, Lindinger S, Daffern SJ, Zito A, Chen LY, Jones LL, Boteva L, Morris DL, Small KS, Fernando MMA, Cunninghame Graham DS, Vyse TJ. Interferon inducible X-linked gene CXorf21 may contribute to sexual dimorphism in Systemic Lupus Erythematosus. Nat Commun, 2019, 10(1): 2164.
doi: 10.1038/s41467-019-10106-2 pmid: 31092820 |
| [106] |
Harris VM, Harley ITW, Kurien BT, Koelsch KA, Scofield RH.Lysosomal pH is regulated in a sex dependent manner in immune cells expressing CXorf21. Front Immunol, 2019, 10: 578.
doi: 10.3389/fimmu.2019.00578 pmid: 31001245 |
| [107] |
Harris VM, Koelsch KA, Kurien BT, Harley ITW, Wren JD, Harley JB, Scofield RH. Characterization of cxorf21 provides molecular insight into female-bias immune response in SLE pathogenesis. Front Immunol, 2019, 10: 2160.
doi: 10.3389/fimmu.2019.02160 pmid: 31695690 |
| [108] |
Heinz LX, Lee J, Kapoor U, Kartnig F, Sedlyarov V, Papakostas K, César-Razquin A, Essletzbichler P, Goldmann U, Stefanovic A, Bigenzahn JW, Scorzoni S, Pizzagalli MD, Bensimon A, Müller AC, King FJ, Li J, Girardi E, Mbow ML, Whitehurst CE, Rebsamen M, Superti-Furga G.TASL is the SLC15A4-associated adaptor for IRF 5 activation by TLR7-9. Nature, 2020, 581(7808): 316-322.
doi: 10.1038/s41586-020-2282-0 |
| [109] |
Bentham J, Morris DL, Graham DSC, Pinder CL, Tombleson P, Behrens TW, Martín J, Fairfax BP, Knight JC, Chen LY, Replogle J, Syvänen AC, Rönnblom L, Graham RR, Wither JE, Rioux JD, Alarcón-Riquelme ME, Vyse TJ. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat Genet, 2015, 47(12): 1457-1464.
doi: 10.1038/ng.3434 pmid: 26502338 |
| [110] | Kwon KS, Cho HY, Chung YJ. Recapitulation of candidate systemic lupus erythematosus-associated variants in Koreans. Genom Inf, 2016, 14: 85-89. |
| [111] | Vawter MP, Harvey PD, DeLisi LE. Dysregulation of X-linked gene expression in Klinefelter’s syndrome and association with verbal cognition. Am J Med Genet B Neuropsychiatr Genet, 2007, 144: 728-734. |
| [112] |
Mackay M, Oswald M, Sanchez-Guerrero J, Lichauco J, Aranow C, Kotkin S, Korsunsky I, Gregersen PK, Diamond B. Molecular signatures in systemic lupus erythematosus: distinction between disease flare and infection. Lupus Sci Med, 2016, 3: e000159.
doi: 10.1136/lupus-2016-000159 |
| [1] | Mao Zhang, Yanyan Wang, Yun Bai, Xuedan Chen, Hong Guo. Investigation on the construction of teaching case base in medical genetics [J]. Hereditas(Beijing), 2024, 46(1): 78-87. |
| [2] | Zhong Bian, Dongping Cao, Wenshu Zhuang, Shuwei Zhang, Qiaoquan Liu, Lin Zhang. Revelation of rice molecular design breeding: the blend of tradition and modernity [J]. Hereditas(Beijing), 2023, 45(9): 718-740. |
| [3] | Xiaoping Lian, Guangfu Huang, Yujiao Zhang, Jing Zhang, Fengyi Hu, Shilai Zhang. The discovery and utilization of favorable genes in Oryza longistaminata [J]. Hereditas(Beijing), 2023, 45(9): 765-780. |
| [4] | Kai Chen, Hao Wang, Yiting Chen, Ke Fu, Zhigang Han, Cong Li, Jinping Si, Donghong Chen. Functional analysis of WOX family genes in Dendrobium catenatum during growth and development [J]. Hereditas(Beijing), 2023, 45(8): 700-714. |
| [5] | Liang Jin, Yujie Chen, Yongjun Chen. Advances in genetic etiology, diagnosis and treatment of developmental and epileptic encephalopathy [J]. Hereditas(Beijing), 2023, 45(7): 553-567. |
| [6] | Jun Ma, Anping Fan, Wusheng Wang, Jinchuan Zhang, Xiaojun Jiang, Ruijun Ma, Sheqiang Jia, Fei Liu, Chuchao Lei, Yongzhen Huang. Analysis of genetic diversity and genetic structure of Qinchuan cattle conservation population using whole-genome resequencing [J]. Hereditas(Beijing), 2023, 45(7): 602-616. |
| [7] | Bingzheng Wang, Chao Zhang, Jiali Zhang, Jin Sun. Conditional editing of the Drosophila melanogaster genome using single transcripts expressing Cas9 and sgRNA [J]. Hereditas(Beijing), 2023, 45(7): 593-601. |
| [8] | Xiao Chen, Jian Lu, Fuqing Yu. Conservation of animal genetic resources in the world and its enlightenment to China [J]. Hereditas(Beijing), 2023, 45(7): 545-552. |
| [9] | Yi Zhang, Zhi-Ying Wu. Pathogenesis and therapeutic advances of cerebral autosomal- dominant arteriopathy with subcortical infarcts and leukoencephalopathy [J]. Hereditas(Beijing), 2023, 45(7): 568-579. |
| [10] | Feifei Li, Yun Wang, Jihai Gu, Yuming Zhang, Fengsong Liu, Zhihua Ni. E2F family play important roles in tumorigenesis [J]. Hereditas(Beijing), 2023, 45(7): 580-592. |
| [11] | Juan Huang, Wenhua Miao, Xiaofeng Guo, Wei Ji. Diagnosis and genetic testing analysis of limb-girdle muscular dystrophy type 2U caused by a compound heterozygous mutation in the ISPD gene [J]. Hereditas(Beijing), 2023, 45(6): 536-542. |
| [12] | Yifan Yu, Zhen OuYang, Juan Guo, Yujun Zhao, Luqi Huang. Progress on regulatory elements of plant plastid genetic engineering [J]. Hereditas(Beijing), 2023, 45(6): 501-513. |
| [13] | Feifei Li, Yanmin Hao, Minlong Cui, Chunlan Piao. Cloning and functional analysis of RADIALIS-like 1 gene from Antirrhinum majus [J]. Hereditas(Beijing), 2023, 45(6): 526-535. |
| [14] | Chenghao Yan, Weiyu Bai, Zhimeng Zhang, Junling Shen, Youjun Wang, Jianwei Sun. The roles and mechanism of STIM1 in tumorigenesis and metastasis [J]. Hereditas(Beijing), 2023, 45(5): 395-408. |
| [15] | Shunze Wang, Feng Jiang, Dongli Zhu, Tie-Lin Yang, Yan Guo. Application of Hi-C technology in three-dimensional genomics research and disease pathogenesis analysis [J]. Hereditas(Beijing), 2023, 45(4): 279-294. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||