Hereditas(Beijing) ›› 2022, Vol. 44 ›› Issue (8): 695-764.doi: 10.16288/j.yczz.22-160
• Research Article • Previous Articles Next Articles
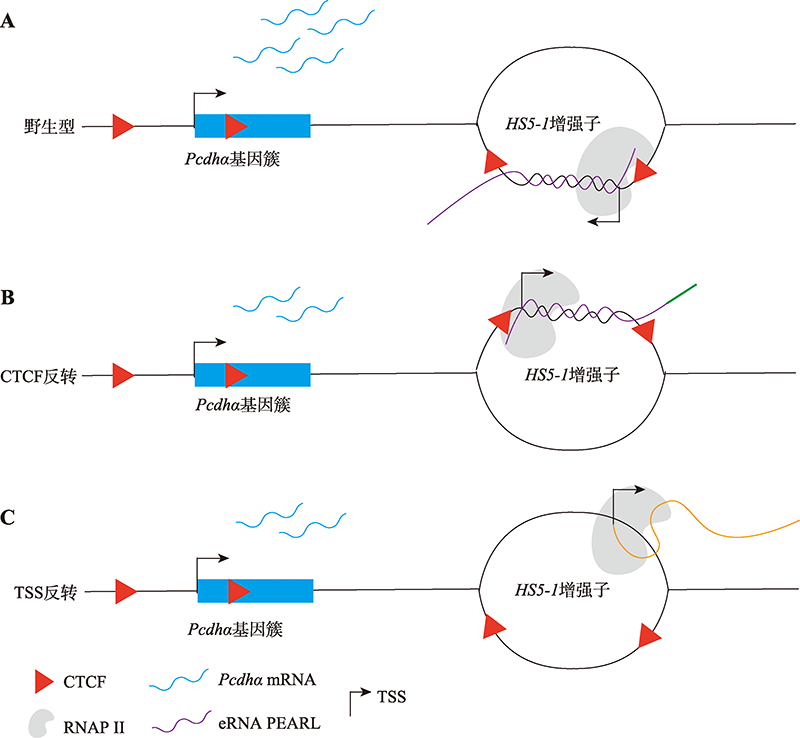
Additional evidence of HS5-1 enhancer eRNA PEARL for protocadherin alpha gene regulation
Siyuan Xu( ), Jia Shou, Qiang Wu(
), Jia Shou, Qiang Wu( )
)
- Center for Comparative Biomedicine, Key laboratory of Systems Biomedicine (Ministry of Education), Institute of Systems Biomedicine, Shanghai Jiao Tong University, Shanghai 200240, China
-
Received:2022-05-15Revised:2022-06-30Online:2022-08-20Published:2022-07-08 -
Contact:Wu Qiang E-mail:xus1yuan@126.com;qiangwu@sjtu.edu.cn -
Supported by:the National Nature Science Foundation of China(31630039);the National Nature Science Foundation of China(91940303);the Science and Technology Commission of Shanghai Municipality Program(19JC1412500);the Science and Technology Commission of Shanghai Municipality Program(21DZ2210200)
Cite this article
Siyuan Xu, Jia Shou, Qiang Wu. Additional evidence of HS5-1 enhancer eRNA PEARL for protocadherin alpha gene regulation[J]. Hereditas(Beijing), 2022, 44(8): 695-764.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1
Primers used in this study"
| 类型 | 引物名称 | 序列(5′→3′) |
|---|---|---|
| PCR | HS51-genetyping-F1 | TTCATCCCCGCTTCCTACTG |
| HS51-genetyping-R1 | CACTCTGATAGTTTATGTATTAGGCTTG | |
| HS51-genetyping-F2 | AGCTGCTGTTTGTGTTTCCGA | |
| HS51-genetyping-R2 | CAGCAAAGGCGGTACAAAAG | |
| qPCR | PEARL-F | AGGCAAAGACACTGGAGTGAAAC |
| PEARL-R | ACCTGTGAACCTTAACTGCCTACTG | |
| Pcdhα-F | AGGAGGCTGGCATTCTACGG | |
| Pcdhα-R | AGGTCCAGCTGTTGCTGTTGAC | |
| GAPDH-F | GGAGTCCACTGGCGTCTTCAC | |
| GAPDH-R | GCAGGAGGCATTGCTGATGAT | |
| sgRNA | HS51-sgRNA1F | ACCGAGAAAGCAATCCATATGGTA |
| HS51-sgRNA1R | AAACTACCATATGGATTGCTTTCT | |
| HS51-sgRNA2F | ACCGGGTGTCTTAGGAAAGCTGAC | |
| HS51-sgRNA2R | AAACGTCAGCTTTCCTAAGACACC | |
| HS51-sgRNA3F | ACCGTGGCTAATTTACAATGCCAG | |
| HS51-sgRNA3R | AAACCTGGCATTGTAAATTAGCCA |
| [1] | Ong CT, Corces VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat Rev Genet, 2011, 12(4): 283-293. |
| [2] |
Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods, 2013, 10(12): 1213-1218.
doi: 10.1038/nmeth.2688 pmid: 24097267 |
| [3] |
Heinz S, Romanoski CE, Benner C, Glass CK. The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol, 2015, 16(3): 144-154.
doi: 10.1038/nrm3949 |
| [4] |
Banerji J, Rusconi S, Schaffner W. Expression of a β-globin gene is enhanced by remote SV40 DNA sequences. Cell, 1981, 27(<W>2 Pt 1):299-308.
pmid: 6277502 |
| [5] |
Andersson R, Sandelin A. Determinants of enhancer and promoter activities of regulatory elements. Nat Rev Genet, 2020, 21(2): 71-87.
doi: 10.1038/s41576-019-0173-8 pmid: 31605096 |
| [6] |
Furey TS. ChIP-seq and beyond: new and improved methodologies to detect and characterize protein-DNA interactions. Nat Rev Genet, 2012, 13(12): 840-852.
doi: 10.1038/nrg3306 |
| [7] |
Korkmaz G, Lopes R, Ugalde AP, Nevedomskaya E, Han RQ, Myacheva K, Zwart W, Elkon R, Agami R.Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9. Nat Biotechnol, 2016, 34(2): 192-198.
doi: 10.1038/nbt.3450 pmid: 26751173 |
| [8] |
Simeonov DR, Gowen BG, Boontanrart M, Roth TL, Gagnon JD, Mumbach MR, Satpathy AT, Lee YJ, Bray NL, Chan AY, Lituiev DS, Nguyen ML, Gate RE, Subramaniam M, Li ZM, Woo JM, Mitros T, Ray GJ, Curie GL, Naddaf N, Chu JS, Ma H, Boyer E, van Gool F, Huang HL, Liu RZ, Tobin VR, Schumann K, Daly MJ, Farh KK, Ansel KM, Ye CJ, Greenleaf WJ, Anderson MS, Bluestone JA, Chang HY, Corn JE, Marson A. Discovery of stimulation-responsive immune enhancers with CRISPR activation. Nature, 2017, 549(7670): 111-115.
doi: 10.1038/nature23875 |
| [9] |
Klann TS, Black JB, Chellappan M, Safi A, Song LY, Hilton IB, Crawford GE, Reddy TE, Gersbach CA. CRISPR-Cas9 epigenome editing enables high-throughput screening for functional regulatory elements in the human genome. Nat Biotechnol, 2017, 35(6): 561-568.
doi: 10.1038/nbt.3853 |
| [10] |
Calo E, Wysocka J.Modification of enhancer chromatin: what, how, and why? Mol Cell, 2013, 49(5): 825-837.
doi: 10.1016/j.molcel.2013.01.038 |
| [11] |
Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao XB, Schmidl C, Suzuki T, Ntini E, Arner E, Valen E, Li K, Schwarzfischer L, Glatz D, Raithel J, Lilje B, Rapin N, Bagger FO, Jørgensen M, Andersen PR, Bertin N, Rackham O, Burroughs AM, Baillie JK, Ishizu Y, Shimizu Y, Furuhata E, Maeda S, Negishi Y, Mungall CJ, Meehan TF, Lassmann T, Itoh M, Kawaji H, Kondo N, Kawai J, Lennartsson A, Daub CO, Heutink P, Hume DA, Jensen TH, Suzuki H, Hayashizaki Y, Müller F, Forrest ARR, Carninci P, Rehli M, Sandelin A. An atlas of active enhancers across human cell types and tissues. Nature, 2014, 507(7493): 455-461.
doi: 10.1038/nature12787 |
| [12] |
Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, Worley PF, Kreiman G, Greenberg ME. Widespread transcription at neuronal activity-regulated enhancers. Nature, 2010, 465(7295): 182-187.
doi: 10.1038/nature09033 |
| [13] |
Pefanis E, Wang JG, Rothschild G, Lim J, Kazadi D, Sun JB, Federation A, Chao J, Elliott O, Liu ZP, Economides AN, Bradner JE, Rabadan R, Basu U. RNA exosome- regulated long non-coding RNA transcription controls super-enhancer activity. Cell, 2015, 161(4): 774-789.
doi: 10.1016/j.cell.2015.04.034 |
| [14] |
Li WB, Notani D, Rosenfeld MG. Enhancers as non- coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet, 2016, 17(4): 207-223.
doi: 10.1038/nrg.2016.4 |
| [15] |
Li X, Fu XD. Chromatin-associated RNAs as facilitators of functional genomic interactions. Nat Rev Genet, 2019, 20(9): 503-519.
doi: 10.1038/s41576-019-0135-1 |
| [16] |
Sartorelli V, Lauberth SM. Enhancer RNAs are an important regulatory layer of the epigenome. Nat Struct Mol Biol, 2020, 27(6): 521-528.
doi: 10.1038/s41594-020-0446-0 pmid: 32514177 |
| [17] |
Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase Ⅱ. Nat Struct Mol Biol, 2007, 14(2): 103-105.
pmid: 17277804 |
| [18] |
Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature, 2013, 494(7438): 497-501.
doi: 10.1038/nature11884 |
| [19] |
Hsieh CL, Fei T, Chen YW, Li TT, Gao YF, Wang XD, Sun T, Sweeney CJ, Lee GSM, Chen SY, Balk SP, Liu XS, Brown M, Kantoff PW. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc Natl Acad Sci USA, 2014, 111(20): 7319-7324.
doi: 10.1073/pnas.1324151111 |
| [20] |
Li WB, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song XY, Oh S, Kim HS, Glass CK, Rosenfeld MG. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature, 2013, 498(7455): 516-520.
doi: 10.1038/nature12210 |
| [21] |
Xiang JF, Yin QF, Chen T, Zhang Y, Zhang XO, Wu Z, Zhang SF, Wang HB, Ge JH, Lu XH, Yang L, Chen LL. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res, 2014, 24(5): 513-531.
doi: 10.1038/cr.2014.35 |
| [22] |
Bose DA, Donahue G, Reinberg D, Shiekhattar R, Bonasio R, Berger SL. RNA binding to CBP stimulates histone acetylation and transcription. Cell, 2017, 168(1-2): 135-149.e22.
doi: 10.1016/j.cell.2016.12.043 |
| [23] |
Dorighi KM, Swigut T, Henriques T, Bhanu NV, Scruggs BS, Nady N, Still CD, Garcia BA, Adelman K, Wysocka J. Mll3 and Mll4 facilitate enhancer RNA synthesis and transcription from promoters independently of H3K4 monomethylation. Mol Cell, 2017, 66(4): 568-576.
doi: 10.1016/j.molcel.2017.04.018 |
| [24] |
Jia ZL, Wu Q. Clustered protocadherins emerge as novel susceptibility loci for mental disorders. Front Neurosci, 2020, 14: 587819.
doi: 10.3389/fnins.2020.587819 |
| [25] |
Wu Q, Jia ZL. Wiring the brain by clustered protocadherin neural codes. Neurosci Bull, 2021, 37(1): 117-131.
doi: 10.1007/s12264-020-00578-4 |
| [26] |
Wu Q, Maniatis T. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell, 1999, 97(6): 779-790.
pmid: 10380929 |
| [27] |
Tasic B, Nabholz CE, Baldwin KK, Kim Y, Rueckert EH, Ribich SA, Cramer P, Wu Q, Axel R, Maniatis T. Promoter choice determines splice site selection in protocadherin α and γ pre-mRNA splicing. Mol Cell, 2002, 10(1): 21-33.
doi: 10.1016/S1097-2765(02)00578-6 |
| [28] |
Wang XZ, Su H, Bradley A. Molecular mechanisms governing Pcdh-γ gene expression: evidence for a multiple promoter and cis-alternative splicing model. Genes Dev, 2002, 16(15): 1890-1905.
doi: 10.1101/gad.1004802 |
| [29] |
Ribich S, Tasic B, Maniatis T. Identification of long-range regulatory elements in the protocadherin-α gene cluster. Proc Natl Acad Sci USA, 2006, 103(52): 19719-19724.
doi: 10.1073/pnas.0609445104 |
| [30] |
Kehayova P, Monahan K, Chen WS, Maniatis T. Regulatory elements required for the activation and repression of the protocadherin-α gene cluster. Proc Natl Acad Sci USA, 2011, 108(41): 17195-17200.
doi: 10.1073/pnas.1114357108 |
| [31] |
Guo Y, Monahan K, Wu HY, Gertz J, Varley KE, Li W, Myers RM, Maniatis T, Wu Q. CTCF/cohesin-mediated DNA looping is required for protocadherin α promoter choice. Proc Natl Acad Sci USA, 2012, 109(51): 21081-21086.
doi: 10.1073/pnas.1219280110 |
| [32] |
Monahan K, Rudnick ND, Kehayova PD, Pauli F, Newberry KM, Myers RM, Maniatis T. Role of CCCTC binding factor (CTCF) and cohesin in the generation of single-cell diversity of Protocadherin-α gene expression. Proc Natl Acad Sci USA, 2012, 109(23): 9125-9130.
doi: 10.1073/pnas.1205074109 |
| [33] | Zhai YN, Xu Q, Guo Y, Wu Q. Characterization of a cluster of CTCF-binding sites in a protocadherin regulatory region. Hereditas(Beijng), 2016, 38(4): 323-336. |
| 翟亚男, 许泉, 郭亚, 吴强. 原钙粘蛋白基因簇调控区域中成簇的CTCF结合位点分析. 遗传, 2016, 38(4): 323-336. | |
| [34] |
Guo Y, Xu Q, Canzio D, Shou J, Li JH, Gorkin DU, Jung I, Wu HY, Zhai YN, Tang YX, Lu YC, Wu YH, Jia ZL, Li W, Zhang MQ, Ren B, Krainer AR, Maniatis T, Wu Q. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell, 2015, 162(4): 900-910.
doi: 10.1016/j.cell.2015.07.038 pmid: 26276636 |
| [35] |
Lu YJ, Shou J, Jia ZL, Wu YH, Li JH, Guo Y, Wu Q. Genetic evidence for asymmetric blocking of higher-order chromatin structure by CTCF/cohesin. Protein Cell, 2019, 10(12): 914-920.
doi: 10.1007/s13238-019-00656-y |
| [36] |
Jia ZL, Li JW, Ge X, Wu YH, Guo Y, Wu Q. Tandem CTCF sites function as insulators to balance spatial chromatin contacts and topological enhancer-promoter selection. Genome Biol, 2020, 21(1): 75.
doi: 10.1186/s13059-020-01984-7 |
| [37] | Wang N, Jia ZL, Wu Q. RFX5 regulates gene expression of the Pcdhα cluster. Hereditas(Beijng), 2020, 42(8): 760-774. |
| 王娜, 甲芝莲, 吴强. RFX5调控原钙粘蛋白α基因簇的表达. 遗传, 2020, 42(8): 760-774. | |
| [38] |
Tang YX, Jia ZL, Xu HL, Da LT, Wu Q. Mechanism of REST/NRSF regulation of clustered protocadherin α genes. Nucleic Acids Res, 2021, 49(8): 4506-4521.
doi: 10.1093/nar/gkab248 |
| [39] |
Canzio D, Nwakeze CL, Horta A, Rajkumar SM, Coffey EL, Duffy EE, Duffié R, Monahan K, O'Keeffe S, Simon MD, Lomvardas S, Maniatis T. Antisense lncRNA transcription mediates DNA demethylation to drive stochastic protocadherin α promoter choice. Cell, 2019, 177(3): 639-653.e15.
doi: 10.1016/j.cell.2019.03.008 |
| [40] |
Zhou YX, Xu SY, Zhang M, Wu Q. Systematic functional characterization of antisense eRNA of protocadherin alpha composite enhancer. Genes Dev, 2021, 35(19-20): 1383-1394.
doi: 10.1101/gad.348621.121 |
| [41] | Li JH, Shou J, Wu Q.DNA fragment editing of genomes by CRISPR/Cas9. Hereditas(Beijng), 2015, 37(10): 992-1002. |
| 李金环, 寿佳, 吴强. CRISPR/Cas9系统在基因组DNA片段编辑中的应用. 遗传, 2015, 37(10): 992-1002. | |
| [42] |
Shou J, Li JH, Liu YB, Wu Q. Precise and predictable CRISPR chromosomal rearrangements reveal principles of Cas9-mediated nucleotide insertion. Mol Cell, 2018, 71(4): 498-509.e4.
doi: S1097-2765(18)30466-0 pmid: 30033371 |
| [43] | Liu PF, Wu Q.Probing 3D genome by CRISPR/Cas9. Hereditas(Beijng), 2020, 42(1): 18-31. |
| 刘沛峰, 吴强. CRISPR/Cas9基因编辑在三维基因组研究中的应用. 遗传, 2020, 42(1): 18-31. | |
| [44] |
Wu Q, Shou J. Toward precise CRISPR DNA fragment editing and predictable 3D genome engineering. J Mol Cell Biol, 2021, 12(11): 828-856.
doi: 10.1093/jmcb/mjaa060 |
| [45] |
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature, 2012, 489(7414): 57-74.
doi: 10.1038/nature11247 |
| [46] |
Gorkin DU, Barozzi I, Zhao Y, Zhang YX, Huang H, Lee AY, Li B, Chiou J, Wildberg A, Ding B, Zhang B, Wang MC, Strattan JS, Davidson JM, Qiu YJ, Afzal V, Akiyama JA, Plajzer-Frick I, Novak CS, Kato M, Garvin TH, Pham QT, Harrington AN, Mannion BJ, Lee EA, Fukuda- Yuzawa Y, He YP, Preissl S, Chee S, Han JY, Williams BA, Trout D, Amrhein H, Yang HB, Cherry JM, Wang W, Gaulton K, Ecker JR, Shen Y, Dickel DE, Visel A, Pennacchio LA, Ren B. An atlas of dynamic chromatin landscapes in mouse fetal development. Nature, 2020, 583(7818): 744-751.
doi: 10.1038/s41586-020-2093-3 |
| [47] |
García-Muse T, Aguiléra A. R loops: from physiological to pathological roles. Cell, 2019, 179(3): 604-618.
doi: S0092-8674(19)31006-2 pmid: 31607512 |
| [48] |
Niehrs C, Luke B. Regulatory R-loops as facilitators of gene expression and genome stability. Nat Rev Mol Cell Biol, 2020, 21(3): 167-178.
doi: 10.1038/s41580-019-0206-3 |
| [49] |
Suzuki ST. Protocadherins and diversity of the cadherin superfamily. J Cell Sci, 1996, 109(Pt 11): 2609-2611.
doi: 10.1242/jcs.109.11.2609 |
| [50] |
Rahnamoun H, Lee J, Sun ZX, Lu HB, Ramsey KM, Komives EA, Lauberth SM. RNAs interact with BRD4 to promote enhanced chromatin engagement and transcription activation. Nat Struct Mol Biol, 2018, 25(8): 687-697.
doi: 10.1038/s41594-018-0102-0 pmid: 30076409 |
| [51] |
Huang ZQ, Liang N, Goñi S, Damdimopoulos A, Wang C, Ballaire R, Jager J, Niskanen H, Han HY, Jakobsson T, Bracken AP, Aouadi M, Venteclef N, Kaikkonen MU, Fan RR, Treuter E. The corepressors GPS2 and SMRT control enhancer and silencer remodeling via eRNA transcription during inflammatory activation of macrophages. Mol Cell, 2021, 81(5): 953-968.e9.
doi: 10.1016/j.molcel.2020.12.040 |
| [1] | Shunze Wang, Feng Jiang, Dongli Zhu, Tie-Lin Yang, Yan Guo. Application of Hi-C technology in three-dimensional genomics research and disease pathogenesis analysis [J]. Hereditas(Beijing), 2023, 45(4): 279-294. |
| [2] | Yan Zhao, Chenxin Wang, Tianming Yang, Chunshuang Li, Lihong Zhang, Dongni Du, Ruoxi Wang, Jing Wang, Min Wei, Xueqing Ba. Linking oxidative DNA lesion 8-OxoG to tumor development and progression [J]. Hereditas(Beijing), 2022, 44(6): 466-477. |
| [3] | Qianbin Zhu, Zhicheng Gan, Xiaocui Li, Yingjie Zhang, Heming Zhao, Xianzhong Huang. Genome-wide identification, phylogenetic and expression of MAPKKK gene family in Arabidopsis pumila [J]. Hereditas(Beijing), 2022, 44(11): 1044-1055. |
| [4] | Yuting Han, Bowen Xu, Yutong Li, Xinyi Lu, Xizhi Dong, Yuhao Qiu, Qinyun Che, Ruibao Zhu, Li Zheng, Xiaochen Li, Xu Si, Jianquan Ni. The cutting edge of gene regulation approaches in model organism Drosophila [J]. Hereditas(Beijing), 2022, 44(1): 3-14. |
| [5] | Cong Zhou, Qiangwei Zhou, Sheng Cheng, Guoliang Li. Research progress of CTCF in mediating 3D genome formation and regulating gene expression [J]. Hereditas(Beijing), 2021, 43(9): 816-821. |
| [6] | Hongyan Lin, Xuan Wang, Cong He, Ziling Zhou, Minkai Yang, Zhongling Wen, Hongwei Han, Guihua Lu, Jinliang Qi, Yonghua Yang. Progress on biosynthesis and function of the natural products of Zi Cao as a traditional Chinese medicinal herb [J]. Hereditas(Beijing), 2021, 43(5): 459-472. |
| [7] | Haidong Xu, Bolin Ning, Fang Mu, Hui Li, Ning Wang. Advances of functional consequences and regulation mechanisms of alternative cleavage and polyadenylation [J]. Hereditas(Beijing), 2021, 43(1): 4-15. |
| [8] | Taotao Wang, Yong Yang, Wei Wei, Chentao Lin, Liuyin Ma. Identification and expression analyses of the NAC transcription factor family in Spartina alterniflora [J]. Hereditas(Beijing), 2020, 42(2): 194-211. |
| [9] | Huiyou Chen, Jianmin Zhang, Baisen Li, Yonglin Deng, Gongwei Zhang. Progress on meiotic gene expression and epigenetic regulation of male sterility in Dzo cattle [J]. Hereditas(Beijing), 2020, 42(11): 1081-1092. |
| [10] | Xiaomeng Gao, Zhihua Zhang. Three-dimensional structure and function of chromatin regulated by “liquid-liquid phase separation” of biological macromolecules. [J]. Hereditas(Beijing), 2020, 42(1): 45-56. |
| [11] | Qichao Yu,Bin Song,Xuanxuan Zou,Ling Wang,Dequan Liu,Bo Li,Kun Ma. Analysis of normal tissues adjacent to the tumour-specific expressed genes in breast cancer [J]. Hereditas(Beijing), 2019, 41(7): 625-633. |
| [12] | Wanjin Xing. The establishment process of lac operon model and the analysis of several teaching problems [J]. Hereditas(Beijing), 2019, 41(6): 548-563. |
| [13] | Tianpei Shi,Li Zhang. Application of whole transcriptomics in animal husbandry [J]. Hereditas(Beijing), 2019, 41(3): 193-205. |
| [14] | Ruoyang Zhao, Yiping Zhao, Bei Li, Gerelchimeg Bou, Xinzhuang Zhang, Togtokh Mongke, Tugeqin Bao, Shurenchimeg Gereliin, Tsimbai Gereltuuin, Chao Li, Dongyi Bai, Manglai Dugarjaviin. Overview of the genetic control of horse coat color patterns [J]. Hereditas(Beijing), 2018, 40(5): 357-368. |
| [15] | Qingqian Ding,Xiaoting Wang,Liqin Hu,Xin Qi,Linhao Ge,Weiya XU,Zhaoshi Xu,Yongbin Zhou,Guanqing Jia,Xianmin Diao,Donghong Min,Youzhi Ma,Ming Chen. MYB-like transcription factor SiMYB42 from foxtail millet (Setaria italica L.) enhances Arabidopsis tolerance to low-nitrogen stress [J]. Hereditas(Beijing), 2018, 40(4): 327-338. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||