遗传 ›› 2020, Vol. 42 ›› Issue (11): 1110-1121.doi: 10.16288/j.yczz.20-213
秦中勇1,2, 石晓1,2, 曹平平1,2, 褚鹰1,2, 管蔚4, 杨楠1,3, 程禾1,2( ), 孙玉洁1,2(
), 孙玉洁1,2( )
)
收稿日期:2020-07-07
修回日期:2020-10-04
出版日期:2020-11-20
发布日期:2020-11-06
通讯作者:
程禾,孙玉洁
E-mail:chenghe@njmu.edu.cn;yujiesun@njmu.edu.cn
作者简介:秦中勇,硕士,专业方向:基因远程调控。E-mail: 基金资助:
Zhongyong Qin1,2, Xiao Shi1,2, Pingping Cao1,2, Ying Chu1,2, Wei Guan4, Nan Yang1,3, He Cheng1,2( ), Yujie Sun1,2(
), Yujie Sun1,2( )
)
Received:2020-07-07
Revised:2020-10-04
Online:2020-11-20
Published:2020-11-06
Contact:
Cheng He,Sun Yujie
E-mail:chenghe@njmu.edu.cn;yujiesun@njmu.edu.cn
Supported by:摘要:
真核生物基因的转录受到近端启动子和远端增强子的共同调控,部分基因的启动子可兼具有增强子的活性。NOXA与BCL2分别是BCL2蛋白家族促凋亡和抗凋亡成员。本课题组前期研究发现,NOXA基因启动子与BCL2基因启动子在染色质三维空间结构上存在相互作用,且NOXA基因启动子区兼有启动子和增强子特征性的组蛋白修饰标记。为进一步探究NOXA启动子是否具有增强子活性、能否在细胞凋亡过程中作为增强子调控BCL2基因表达,本研究利用染色质构象捕获(chromosome conformation capture, 3C)、实时荧光定量PCR (quantitative real-time PCR, qRT-PCR)和荧光素酶报告基因等检测技术在喜树碱诱导的MCF-7细胞凋亡模型中证实,NOXA启动子兼具增强子活性,并可通过形成染色质环结构远程调控BCL2基因表达。NOXA启动子的调控属性与凋亡信号强弱密切相关,在较弱凋亡信号刺激下(1 μmol/L喜树碱处理),NOXA启动子主要发挥增强子功能;随着凋亡刺激信号的加强(10 μmol/L喜树碱处理),NOXA启动子活性增强,主要调控其基因自身的表达,促进细胞凋亡。染色质免疫共沉淀(chromatin immunoprecipitation, ChIP)证实NOXA启动子区启动子活性和增强子活性的动态变化与其组蛋白修饰标志一致。本研究为进一步探讨BCL2家族成员对细胞凋亡刺激做出协同反应的机制提供了新的线索。
秦中勇, 石晓, 曹平平, 褚鹰, 管蔚, 杨楠, 程禾, 孙玉洁. 细胞凋亡反应中NOXA基因启动子发挥增强子功能调节BCL2基因表达[J]. 遗传, 2020, 42(11): 1110-1121.
Zhongyong Qin, Xiao Shi, Pingping Cao, Ying Chu, Wei Guan, Nan Yang, He Cheng, Yujie Sun. The NOXA promoter could function as an active enhancer to regulate the expression of BCL2 in the apoptosis response[J]. Hereditas(Beijing), 2020, 42(11): 1110-1121.
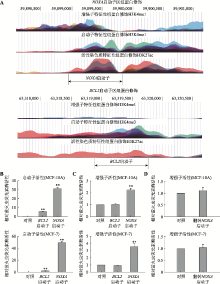
图1
分析和鉴定NOXA基因启动子的增强子活性 A:UCSC数据库数据显示的NOXA基因启动子区(上图)和BCL2基因启动子区(下图)的组蛋白H3K4me1、H3K4me3、H3K27ac修饰特征。B:启动子活性验证。将NOXA启动子和BCL2启动子分别插入至pGL3-basic荧光素酶基因上游后转染进MCF-10A、MCF-7细胞中(对照为pGL3-basic空载质粒),验证二者的启动子活性;C:增强子活性验证。将NOXA启动子和BCL2启动子分别插入至pGL3-promoter荧光素酶基因下游后转染MCF-10A和MCF-7细胞(对照为pGL3-promoter空载质粒),检测二者的增强子活性,结果显示NOXA启动子兼具有增强子活性。D:翻转NOXA启动子后增强子活性验证。将NOXA启动子翻转后插入至pGL3-promoter荧光素酶基因下游后转染MCF-10A和MCF-7细胞(对照为pGL3-promoter空载质粒),检测NOXA启动子翻转后的增强子活性。*:P<0.05;**:P<0.01。"
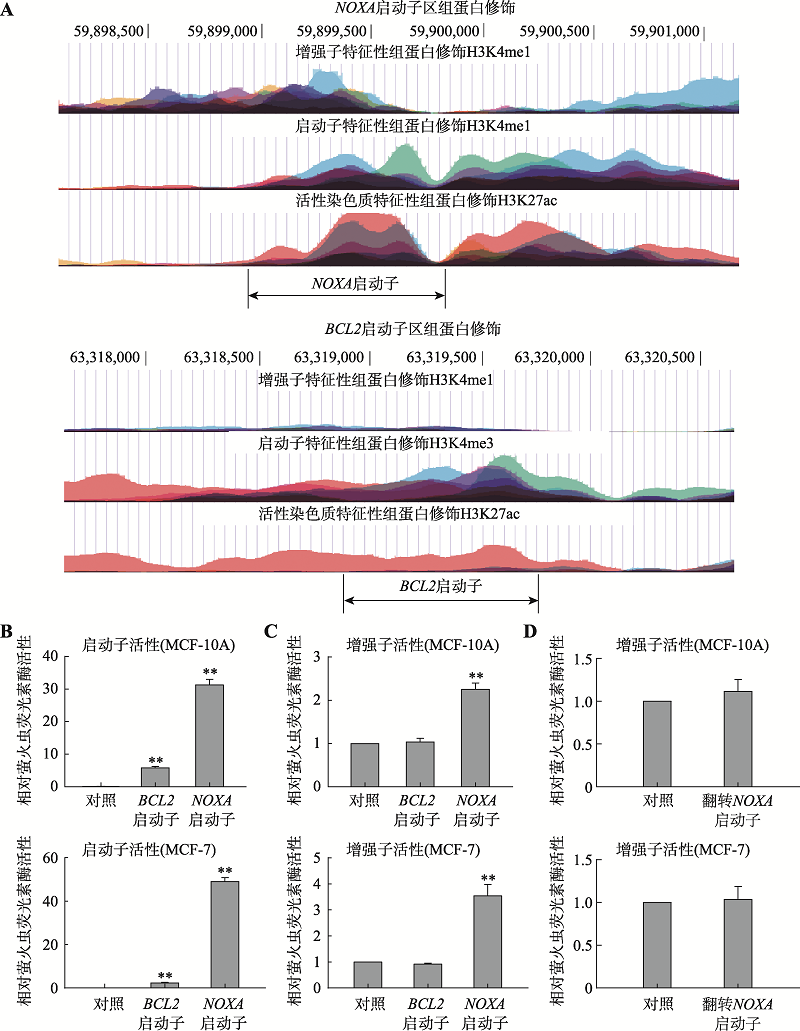
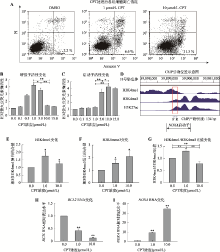
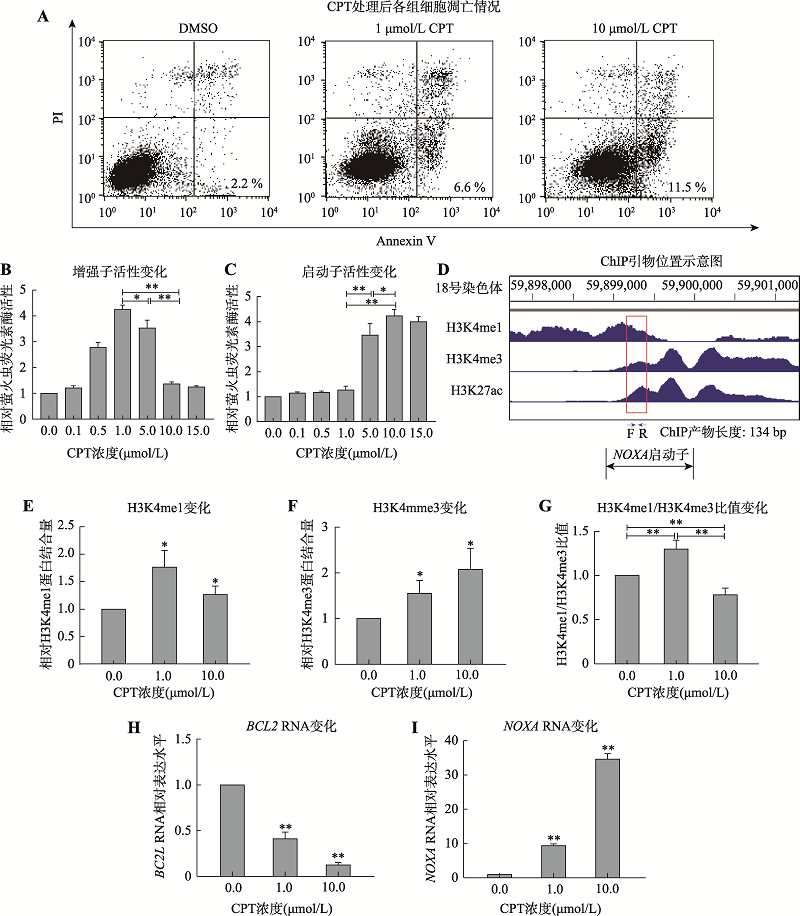
图4
喜树碱诱导NOXA基因启动子向增强子转变 A:喜树碱处理后各组凋亡情况。分别以低浓度(1μmol/L)和高浓度(10μmol/L)喜树碱处理MCF-7细胞10 h,收获细胞后运用流式细胞术测定细胞凋亡。B和C:利用报告基因系统分别检测不同浓度喜树碱处理MCF-7细胞后10 h后,NOXA基因启动子和增强子活性变化。D:ENCODE数据库中的MCF-7细胞H3K4me1和H3K4me3修饰的ChIP-seq数据。红框圈出的部分为H3K4me1和H3K4me3组蛋白修饰重叠区域,在该部分设计合适的ChIP引物。E和F:利用ChIP验证不同浓度喜树碱处理MCF-7细胞后NOXA启动子区组蛋白修饰H3K4me1和H3K4me3的变化。G: ChIP数据H3K4me1/H3K4me3的比值变化。H:喜树碱处理MCF-7细胞BCL2基因的mRNA水平变化。I:喜树碱处理MCF-7细胞NOXA基因的mRNA水平变化。*:P<0.05;**:P<0.01。"
| [1] |
Kim TK, Shiekhattar R . Architectural and functional commonalities between enhancers and promoters. Cell, 2015,162(5):948-959.
doi: 10.1016/j.cell.2015.08.008 pmid: 26317464 |
| [2] |
Khoury G, Gruss P . Enhancer elements. Cell, 1983,33(2):313-314.
doi: 10.1016/0092-8674(83)90410-5 pmid: 6305503 |
| [3] |
Butler JEF, Kadonaga JT . Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev, 2001,15(19):2515-2519.
doi: 10.1101/gad.924301 pmid: 11581157 |
| [4] |
Qi HY, Zhang ZJ, Li YJ, Fang XD . Role of chromatin conformation in eukaryotic gene regulation. Hereditas (Beijing), 2011,33(12):1291-1299.
doi: 10.3724/SP.J.1005.2011.01291 |
|
亓合媛, 张昭军, 李雅娟, 方向东 . 染色质构象调控真核基因的表达. 遗传, 2011,33(12):1291-1299.
doi: 10.3724/SP.J.1005.2011.01291 |
|
| [5] |
Fromm M, Berg P . Deletion mapping of DNA regions required for SV40 early region promoter function in vivo. J Mol Appl Genet, 1982,1(5):457-481.
pmid: 6296253 |
| [6] |
Smale ST, Kadonaga JT . The RNA polymerase II core promoter. Annu Rev Biochem, 2003,72:449-479.
doi: 10.1146/annurev.biochem.72.121801.161520 pmid: 12651739 |
| [7] |
Atchison ML . Enhancers: mechanisms of action and cell specificity. Annu Rev Cell Biol, 1988,4:127-153.
doi: 10.1146/annurev.cb.04.110188.001015 pmid: 2848550 |
| [8] |
Morris JR, Petrov, DA, Lee AM, Wu CT, . Enhancer choice in cis and in trans in Drosophila melanogaster: role of the promoter. Genetics, 2004,167(4):1739-1747.
doi: 10.1534/genetics.104.026955 pmid: 15342512 |
| [9] |
He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang QB, Zhang Y, Xu KX, Ni M, Lupien M, Mieczkowski P, Lieb JD, Zhao KJ, Brown M, Liu XS . Nucleosome dynamics define transcriptional enhancers. Nat Genet, 2010,42(4):343-347.
doi: 10.1038/ng.545 pmid: 20208536 |
| [10] |
Mikhaylichenko O, Bondarenko V, Harnett D, Schor IE, Males M, Viales RR, Furlong EEM . The degree of enhancer or promoter activity is reflected by the levels and directionality of eRNA transcription. Genes Dev, 2018,32(1):42-57.
doi: 10.1101/gad.308619.117 pmid: 29378788 |
| [11] |
Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao XB, Schmidl C, Suzuki T, Ntini E, Arner E, Valen E, Li K, Schwarzfischer L, Glatz D, Raithel J, Lilje B, Rapin N, Bagger FO, Jørgensen M, Andersen PR, Bertin N, Rackham O, Burroughs AM, Baillie JK, Ishizu Y, Shimizu Y, Furuhata E, Maeda S, Negishi Y, Mungall CJ, Meehan TF, Lassmann T, Itoh M, Kawaji H, Kondo N, Kawai J, Lennartsson A, Daub CO, Heutink P, Hume DA, Jensen TH, Suzuki H, Hayashizaki Y, Müller F, Forrest ARR, Carninci P, Rehli M, Sandelin A . An atlas of active enhancers across human cell types and tissues. Nature, 2014,507(7493):455-461.
doi: 10.1038/nature12787 |
| [12] |
Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang XM, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B . Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature, 2009,459(7243):108-112.
doi: 10.1038/nature07829 pmid: 19295514 |
| [13] |
Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, JaenischS R. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA, 2010,107(50):21931-21936.
doi: 10.1073/pnas.1016071107 pmid: 21106759 |
| [14] |
Charlet J, Duymich CE, Lay FD, Mundbjerg K, Dalsgaard Sorensen K, Liang GN, Jones PA . Bivalent regions of cytosine methylation and H3K27 acetylation suggest an active role for DNA methylation at enhancers. Mol Cell, 2016,62(3):422-431.
doi: 10.1016/j.molcel.2016.03.033 pmid: 27153539 |
| [15] |
Local A, Huang H, Albuquerque CP, Singh N, Lee AY, Wang W, Wang CC, Hsia JE, Shiau AK, Ge K, Corbett KD, Wang D, Zhou HL, Ren B . Identification of H3K4me1- associated proteins at mammalian enhancers. Nat Genet, 2018,50(1):73-82.
doi: 10.1038/s41588-017-0015-6 pmid: 29255264 |
| [16] |
Dao LTM, Galindo-Albarrán AO, Castro-Mondragon JA, Andrieu-Soler C, Medina-Rivera A, Souaid C, Charbonnier G, Griffon A, Vanhille L, Stephen T, Alomairi J, Martin D, Torres M, Fernandez N, Soler E, van Helden J, Puthier D, Spicuglia S, . Genome-wide characterization of mammalian promoters with distal enhancer functions. Nat Genet, 2017,49(7):1073-1081.
doi: 10.1038/ng.3884 pmid: 28581502 |
| [17] |
Hua JT, Ahmed M, Guo HY, Zhang YZ, Chen SJ, Soares F, Lu J, Zhou S, Wang M, Li H, Larson NB, McDonnell SK, Patel PS, Liang Y, Yao CQ, van der Kwast T, Lupien M, Feng FY, Zoubeidi A, Tsao MS, Thibodeau SN, Boutros PC, He HH. Risk SNP-Mediated promoter-enhancer switching drives prostate cancer through lncRNA PCAT19. Cell, 2018, 174(3): 564-575.e518.
doi: 10.1016/j.cell.2018.06.014 pmid: 30033362 |
| [18] |
Medina-Rivera A, Santiago-Algarra D, Puthier D, Spicuglia S . Widespread enhancer activity from core promoters. Trends Biochem Sci, 2018,43(6):452-468.
doi: 10.1016/j.tibs.2018.03.004 pmid: 29673772 |
| [19] |
Zinkel S, Gross A, Yang E . BCL2 family in DNA damage and cell cycle control. Cell Death Differ, 2006,13(8):1351-1359.
doi: 10.1038/sj.cdd.4401987 pmid: 16763616 |
| [20] |
Hwang KT, Kim K, Chang JH, Oh S, Kim YA, Lee JY, Jung SH, Choi IS . BCL2 regulation according to molecular subtype of breast cancer by analysis of the cancer genome atlas database. Cancer Res Treat, 2018,50(3):658-669.
doi: 10.4143/crt.2017.134 pmid: 28701032 |
| [21] |
Radha G, Raghavan SC . BCL2: A promising cancer therapeutic target. Biochim Biophys Acta Rev Cancer, 2017,1868(1):309-314.
doi: 10.1016/j.bbcan.2017.06.004 pmid: 28647470 |
| [22] |
Lisachev PD, Pustyl'nyak VO, Shtark MB, . Expression of Bcl2 family genes in the early phase of long-term potentiation. Bull Exp Biol Med, 2014,158(1):77-79.
doi: 10.1007/s10517-014-2696-5 pmid: 25403402 |
| [23] |
Moldoveanu T, Follis AV, Kriwacki RW, Green DR . Many players in BCL-2 family affairs. Trends Biochem Sci, 2014,39(3):101-111.
doi: 10.1016/j.tibs.2013.12.006 pmid: 24503222 |
| [24] |
Kim JY, Ahn HJ, Ryu JH, Suk K, Park JH . BH3-only protein Noxa is a mediator of hypoxic cell death induced by hypoxia-inducible factor 1α. J Exp Med, 2004,199(1):113-124.
doi: 10.1084/jem.20030613 pmid: 14699081 |
| [25] |
Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, Colman PM, Day CL, Adams JM, Huang DC . Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell, 2005,17(3):393-403.
doi: 10.1016/j.molcel.2004.12.030 pmid: 15694340 |
| [26] |
Yang Y, Wang ZD, Sun L, Shao LP, Yang N, Yu DW, Zhang X, Han X, Sun YJ . SATB1 mediates long-range chromatin interactions: a dual regulator of anti-apoptotic BCL2 and Pro-apoptotic NOXA genes. PLoS One, 2015,10(9):e0139170.
doi: 10.1371/journal.pone.0139170 pmid: 26422397 |
| [27] |
Zhang JJ, Ma CY, Han X, Durrin LK, Sun YJ . The bcl-2 major breakpoint region (mbr) possesses transcriptional regulatory function. Gene, 2006,379:127-131.
doi: 10.1016/j.gene.2006.05.002 pmid: 16777355 |
| [28] |
Ma C, Zhang J, Durrin LK, Lv J, Zhu D, Han X, Sun Y . The BCL2 major breakpoint region (mbr) regulates gene expression. Oncogene, 2007,26(18):2649-2657.
doi: 10.1038/sj.onc.1210069 pmid: 17057736 |
| [29] |
Gong FR, Sun L, Wang ZD, Shi JF, Li W, Wang SM, Han X, Sun YJ . The BCL2 gene is regulated by a special AT-rich sequence binding protein 1-mediated long range chromosomal interaction between the promoter and the distal element located within the 3'-UTR. Nucleic Acids Res, 2011,39(11):4640-4652.
doi: 10.1093/nar/gkr023 pmid: 21310710 |
| [30] | Hu WJ, Sun YJ . The mbr-FPGS efficient expression plasmid enhances the sensitivity of Jurkat cells to methotrexate. Hereditas(Beijing), 2012,34(6):705-710. |
| 胡文佳, 孙玉洁 . mbr-FPGS高效表达质粒增强Jurkat细胞对甲氨蝶呤的敏感性. 遗传, 2012,34(6):705-710. | |
| [31] |
Zweig AS, Karolchik D, Kuhn RM, Haussler D, Kent WJ . UCSC genome browser tutorial. Genomics, 2008,92(2):75-84.
doi: 10.1016/j.ygeno.2008.02.003 |
| [32] | Zhao JC, Chai Z, Guo SM, Liu ZH . Analysis of SOX2 gene promoter activity in porcine early embryonic development. Hereditas(Beijing), 2019,41(10):950-961. |
| 赵剑超, 柴壮, 郭诗萌, 刘忠华 . 猪早期胚胎发育中SOX2基因启动子活性分析. 遗传, 2019,41(10):950-961. | |
| [33] |
Li GL, Ruan XN, Auerbach RK, Sandhu KS, Zheng MZ, Wang P, Poh HM, Goh Y, Lim J, Zhang JY, Sim HS, Peh SQ, Mulawadi FH, Ong CT, Orlov YL, Hong SZ, Zhang ZZ, Landt S, Raha D, Euskirchen G, Wei CL, Ge WH, Wang HE, Davis C, Fisher-Aylor KI, Mortazavi A, Gerstein M, Gingeras T, Wold B, Sun Y, Fullwood MJ, Cheung E, Liu E, Sung WK, Snyder M, Ruan YJ . Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell, 2012,148(1-2):84-98.
doi: 10.1016/j.cell.2011.12.014 |
| [34] |
Dekker J, Rippe K, Dekker M, Kleckner N . Capturing chromosome conformation. Science, 2002,295(5558):1306-1311.
doi: 10.1126/science.1067799 pmid: 11847345 |
| [35] |
Cope NF, Fraser P, . Chromosome conformation capture. Cold Spring Harb Protoc, 2009, 2009(2): pdb.prot5137.
doi: 10.1101/pdb.top098210 pmid: 29438064 |
| [36] |
Sati S, Cavalli G . Chromosome conformation capture technologies and their impact in understanding genome function. Chromosoma, 2017,126(1):33-44.
doi: 10.1007/s00412-016-0593-6 pmid: 27130552 |
| [37] |
Orekhova AS, Rubtsov PM . Bidirectional promoters in the transcription of mammalian genomes. Biochemistry (Mosc), 2013,78(4):335-341.
doi: 10.1134/S0006297913040020 |
| [38] |
Scruggs BS, Gilchrist DA, Nechaev S, Muse GW, Burkholder A, Fargo DC, Adelman K . Bidirectional transcription arises from two distinct hubs of transcription factor binding and active chromatin. Mol Cell, 2015,58(6):1101-1112.
doi: 10.1016/j.molcel.2015.04.006 pmid: 26028540 |
| [39] |
Nejepinska J, Malik R, Moravec M, Svoboda P . Deep sequencing reveals complex spurious transcription from transiently transfected plasmids. PLoS One, 2012,7(8):e43283.
doi: 10.1371/journal.pone.0043283 pmid: 22916237 |
| [40] |
Li QY, Zu YG, Shi RZ, Yao LP . Review camptothecin: current perspectives. Curr Med Chem, 2006,13(17):2021-2039.
doi: 10.2174/092986706777585004 pmid: 16842195 |
| [41] |
Sriram D, Yogeeswari P, Thirumurugan R, Bal TR . Camptothecin and its analogues: a review on their chemotherapeutic potential. Nat Prod Res, 2005,19(4):393-412.
doi: 10.1080/14786410412331299005 pmid: 15938148 |
| [42] |
Pommier Y . Topoisomerase I inhibitors: camptothecins and beyond. Nat Rev Cancer, 2006,6(10):789-802.
doi: 10.1038/nrc1977 pmid: 16990856 |
| [43] | Ding N, Qu HZ, Fang XD . The ENCODE project and functional genomics studies. Hereditas(Beijing), 2014,36(3):237-247. |
| 丁楠, 渠鸿竹, 方向东 . ENCODE计划和功能基因组研究. 遗传, 2014,36(3):237-247. | |
| [44] |
Campbell KJ, Tait SWG . Targeting BCL-2 regulated apoptosis in cancer. Open Biol, 2018,8(5):180002.
doi: 10.1098/rsob.180002 pmid: 29769323 |
| [45] |
Oing C, Tennstedt P, Simon R, Volquardsen J, Borgmann K, Bokemeyer C, Petersen C, Dikomey E, Rothkamm K, Mansour WY . BCL2-overexpressing prostate cancer cells rely on PARP1-dependent end-joining and are sensitive to combined PARP inhibitor and radiation therapy. Cancer Lett, 2018,423:60-70.
doi: 10.1016/j.canlet.2018.03.007 pmid: 29526801 |
| [46] |
Diao YR, Fang RX, Li B, Meng ZP, Yu JT, Qiu YJ, Lin KC, Huang H, Liu T, Marina RJ, Jung I, Shen Y, Guan KL, Ren B . A tiling-deletion-based genetic screen for cis-regulatory element identification in mammalian cells. Nat Methods, 2017,14(6):629-635.
pmid: 28417999 |
| [47] | Mumbach MR, Satpathy AT, Boyle EA, Dai C, Gowen BG, Cho SW, Nguyen ML, Rubin AJ, Granja JM, Kazane KR, Wei YN, Nguyen T, Greenside PG, Corces MR, Tycko J, Simeonov DR, Suliman N, Li R, Xu J, Flynn RA, Kundaje A, Khavari PA, Marson A, Corn JE, Quertermous T, Greenleaf WJ, Chang HY . Enhancer connectome in primary human cells identifies target genes of disease- associated DNA elements. Nat Genet, 2017,49(11):1602-1612. |
| [48] |
Weintraub AS, Li CH, Zamudio AV, Sigova AA, Hannett NM, Day DS, Abraham BJ, Cohen MA, Nabet B, Buckley DL, Guo YE, Hnisz D, Jaenisch R, Bradner JE, Gray NS, Young RA . YY1 Is a structural regulator of enhancer- promoter loops. Cell, 2017, 171(7): 1573-1588.e1528.
pmid: 29224777 |
| [1] | 陈秀丽, 黄海燕, 吴强. 靶向敲除β-珠蛋白基因座控制区增强子HS2对K562细胞转录组的影响[J]. 遗传, 2022, 44(9): 783-797. |
| [2] | 徐思远, 寿佳, 吴强. HS5-1增强子eRNA PEARL对原钙粘蛋白α基因簇的表达调控[J]. 遗传, 2022, 44(8): 695-764. |
| [3] | 漆思晗, 王棨临, 张俊有, 刘倩, 李春燕. 增强子调控癌症发生发展的机制研究[J]. 遗传, 2022, 44(4): 275-288. |
| [4] | 王翠玲, 刘信燚, 王亚会, 张争, 王治东, 周钢桥. MCM2通过抑制p53信号通路促进胆管癌细胞的增殖、迁移和侵袭[J]. 遗传, 2022, 44(3): 230-244. |
| [5] | 万星琦, 魏婉珍, 郭胜良, 崔一笑, 景雪莹, 黄露杰, 马捷. BMP2基因远程调控元件的功能分析[J]. 遗传, 2022, 44(12): 1141-1147. |
| [6] | 王玲, 李金环, 黄海燕, 吴强. 串联反向CTCF位点的系列删除揭示增强子调控HOXD基因簇表达的平衡[J]. 遗传, 2021, 43(8): 775-791. |
| [7] | 刘倩, 李春燕. 增强子的鉴定及其在肿瘤研究中的应用[J]. 遗传, 2020, 42(9): 817-831. |
| [8] | 余同露,蔡栋梁,朱根凤,叶晓娟,闵太善,陈红岩,卢大儒,陈浩明. CSN4基因干扰对乳腺癌MDA-MB-231细胞增殖和凋亡的影响[J]. 遗传, 2019, 41(4): 318-326. |
| [9] | 吴志强, 米泽云. 超级增强子在肿瘤研究中的进展[J]. 遗传, 2019, 41(1): 41-51. |
| [10] | 程霄,杨琼,谭镇东,谭娅,蒲红州,赵雪,张顺华,朱砺. 增强子RNA研究现状[J]. 遗传, 2017, 39(9): 784-797. |
| [11] | 李俊涛,赵薇,李丹丹,冯静,巴贵,宋天增,张红平. miR-101a靶向EZH2促进山羊骨骼肌卫星细胞的分化[J]. 遗传, 2017, 39(9): 828-836. |
| [12] | 孙长斌, 张曦. 超级增强子研究进展[J]. 遗传, 2016, 38(12): 1056-1068. |
| [13] | 赵建元,丁寄葳,米泽云,周金明,魏涛,岑山. HIV-1初始传播病毒Vpr基因遗传变异对诱导G2期阻滞及细胞凋亡的影响[J]. 遗传, 2015, 37(5): 480-486. |
| [14] | 毕丹,徐扬,逄越,李庆伟. 质膜组分磷脂酰丝氨酸外翻的分子调控机制[J]. 遗传, 2015, 37(2): 140-147. |
| [15] | 张蕾, 隋御, 王婷, 李利坚, 李元杰, 金彩霞, 徐方. hMMS2基因对结肠癌细胞耐药逆转的影响[J]. 遗传, 2014, 36(4): 346-353. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||

www.chinagene.cn
备案号:京ICP备09063187号-4
总访问:,今日访问:,当前在线:
