遗传 ›› 2022, Vol. 44 ›› Issue (8): 695-764.doi: 10.16288/j.yczz.22-160
收稿日期:2022-05-15
修回日期:2022-06-30
出版日期:2022-08-20
发布日期:2022-07-08
作者简介:徐思远,在读硕士研究生,专业方向:生物学。E-mail: 基金资助:
Siyuan Xu( ), Jia Shou, Qiang Wu(
), Jia Shou, Qiang Wu( )
)
Received:2022-05-15
Revised:2022-06-30
Published:2022-08-20
Online:2022-07-08
Supported by:摘要:
远端增强子对关键靶基因的表达调控通常可以决定细胞的命运和功能,激活的增强子可以双向转录产生长非编码(long noncoding)增强子RNA (enhancer RNA, eRNA)调控靶基因表达,课题组前期研究发现增强子eRNA能够通过形成R环(R-loop)来促进增强子与靶基因的染色质远距离互作,引起局部三维基因组TAD (topologically associated domain)的改变。为了进一步探究eRNA在基因转录过程中的生物学功能,本研究选取原钙粘蛋白(protocadherin, Pcdh)基因簇的增强子eRNA PEARL (Pcdh eRNA associated with R-loop formation)作为研究对象,通过CRISPR (clustered regularly interspaced short palindromic repeats) DNA片段编辑技术、逆转录PCR、荧光定量PCR等遗传学和分子生物学实验,揭示了增强子eRNA PEARL对Pcdhα基因簇表达的促进作用。首先,本研究通过分析不同组织中HS5-1增强子eRNA发现其表达具有组织特异性;其次,通过CRISPR诱导HS5-1增强子内CTCF结合位点的反转或缺失会造成增强子eRNA PEARL转录水平降低至2%~10%,同时Pcdhα基因簇的转录水平也会降低至原来的13%~68%;最后,利用CRISPR DNA片段编辑技术删除HS5-1 eRNA双向转录起始位点或反转eRNA PEARL的转录起始位点后,Pcdhα基因簇的转录水平下降了约60%或40%,表明HS5-1增强子eRNA在调控基因表达中发挥重要作用。以上研究结果进一步证实了HS5-1增强子的转录产物可以调控Pcdhα基因簇的表达,为后续研究原钙粘蛋白基因簇在大脑中的表达调控机制提供了新的思路或方向。
徐思远, 寿佳, 吴强. HS5-1增强子eRNA PEARL对原钙粘蛋白α基因簇的表达调控[J]. 遗传, 2022, 44(8): 695-764.
Siyuan Xu, Jia Shou, Qiang Wu. Additional evidence of HS5-1 enhancer eRNA PEARL for protocadherin alpha gene regulation[J]. Hereditas(Beijing), 2022, 44(8): 695-764.
表1
引物序列"
| 类型 | 引物名称 | 序列(5′→3′) |
|---|---|---|
| PCR | HS51-genetyping-F1 | TTCATCCCCGCTTCCTACTG |
| HS51-genetyping-R1 | CACTCTGATAGTTTATGTATTAGGCTTG | |
| HS51-genetyping-F2 | AGCTGCTGTTTGTGTTTCCGA | |
| HS51-genetyping-R2 | CAGCAAAGGCGGTACAAAAG | |
| qPCR | PEARL-F | AGGCAAAGACACTGGAGTGAAAC |
| PEARL-R | ACCTGTGAACCTTAACTGCCTACTG | |
| Pcdhα-F | AGGAGGCTGGCATTCTACGG | |
| Pcdhα-R | AGGTCCAGCTGTTGCTGTTGAC | |
| GAPDH-F | GGAGTCCACTGGCGTCTTCAC | |
| GAPDH-R | GCAGGAGGCATTGCTGATGAT | |
| sgRNA | HS51-sgRNA1F | ACCGAGAAAGCAATCCATATGGTA |
| HS51-sgRNA1R | AAACTACCATATGGATTGCTTTCT | |
| HS51-sgRNA2F | ACCGGGTGTCTTAGGAAAGCTGAC | |
| HS51-sgRNA2R | AAACGTCAGCTTTCCTAAGACACC | |
| HS51-sgRNA3F | ACCGTGGCTAATTTACAATGCCAG | |
| HS51-sgRNA3R | AAACCTGGCATTGTAAATTAGCCA |
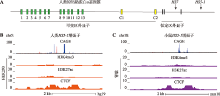
图1
原钙粘蛋白α基因簇HS5-1 eRNA的组织特异性分析 A:人类原钙粘蛋白α基因簇(Pcdhα)示意图。人类Pcdhα基因簇由15个可变区外显子和3个恒定区外显子组成,15个可变区外显子包括13个随机表达型外显子和2个C型外显子。每个Pcdhα基因的可变区外显子都携带自己的启动子,通过顺式剪接与下游3个恒定区外显子相连。B:ENCODE数据库中[45]人类HEK293细胞系中HS5-1增强子处eRNA的特异性分析。依次为H3K4me3、H3K27ac和CTCF的ChIP-seq结果(橘色)。C:ENCODE数据库[46]小鼠肾脏组织(紫色)中HS5-1增强子处eRNA的组织特异性分析。"
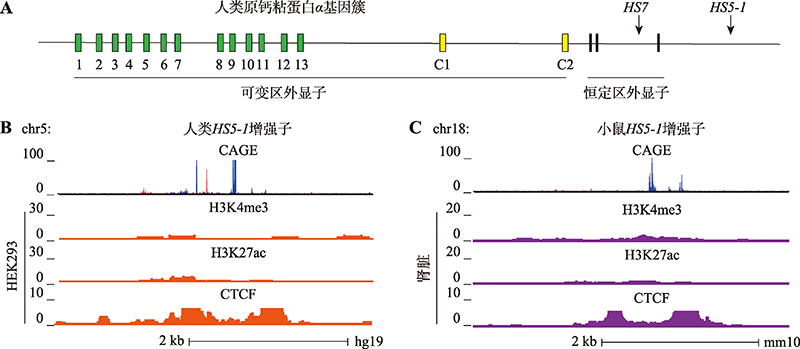
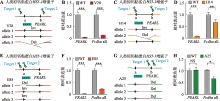
图2
HS5-1增强子CTCF位点方向改变导致eRNA转录水平降低 A:HS5-1增强子CBS反转的单克隆细胞株基因型示意图。B:增强子eRNA PEARL和Pcdhα基因表达的定量分析。C:HS5-1增强子CBS编辑后的单克隆细胞株基因型示意图。D:增强子eRNA PEARL和Pcdhα基因表达的定量结果。E:HS5-1增强子CBSa反转的单克隆细胞株基因型示意图。F:增强子eRNA PEARL和Pcdhα基因表达的定量分析。G:HS5-1增强子CBSb删除的单克隆细胞株基因型示意图。H:增强子eRNA PEARL和Pcdhα基因表达的定量结果。NS:没有显著性差异;*:P<0.05;**:P<0.01;***:P<0.001。"
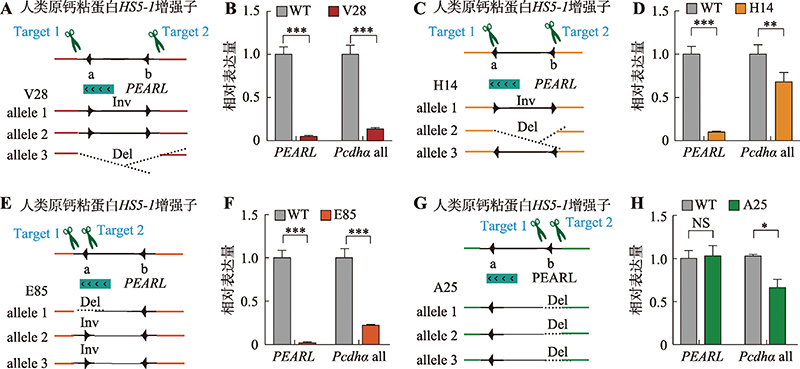
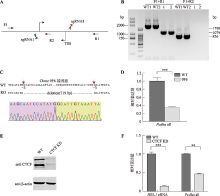
图3
使用CRISPR DNA片段编辑技术删除HS5-1增强子eRNA双向转录起始位点 A:敲除HS5-1增强子eRNA转录起始区所用sgRNAs及单克隆基因型鉴定所用引物示意图。B:单克隆细胞株基因型鉴定凝胶电泳图。以野生型细胞作为对照,F1和R1的目的产物片段为1798 bp,删除转录起始区后产物片段为1079 bp,F1和R2的目的产物片段为826 bp。C:单克隆细胞株基因型鉴定一代测序图。红色碱基为PAM位点,红色箭头表示Cas9切割位点;粉色框内为上游接口处碱基,黄色框内为下游接口处碱基。D:野生型克隆、HS5-1增强子eRNA转录起始区域删除的单克隆细胞株中Pcdhα基因表达的定量分析。E:敲低CTCF后的蛋白质免疫印记。F:CTCF敲低后的HS5-1增强子eRNA和Pcdhα基因表达的定量分析。TSS:转录起始位点;WT:野生型;KD:敲低。**:P<0.01;***:P<0.001。"
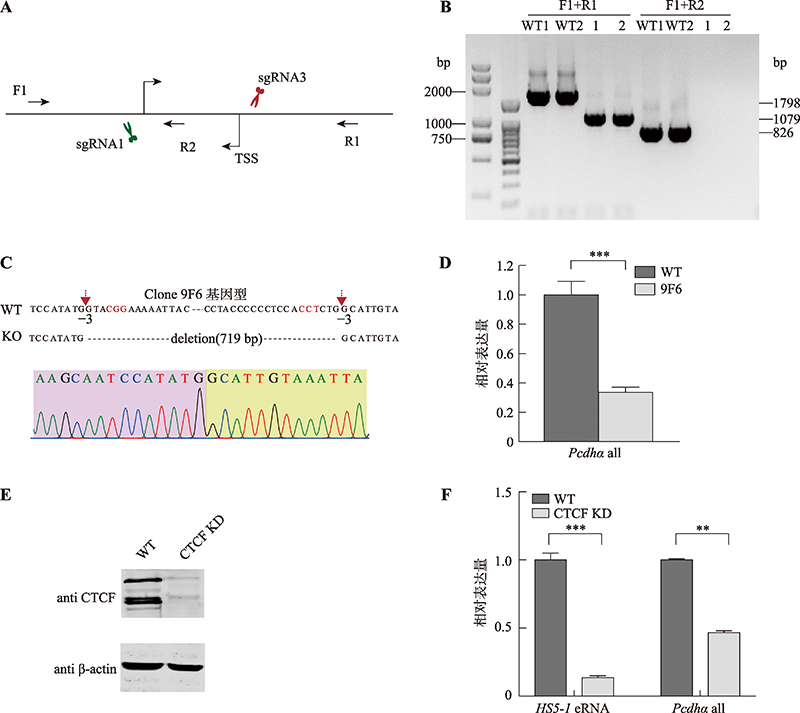
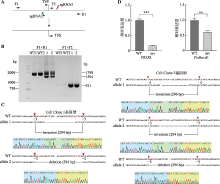
图4
使用CRISPR DNA片段编辑技术反转HS5-1增强子eRNA PEARL的转录起始位点 A:反转eRNA PEARL转录起始位点所用sgRNAs及单克隆基因型鉴定所用引物示意图。B:单克隆细胞株基因型鉴定凝胶电泳图。以野生型细胞作为对照,F1和R1的目的产物片段为1798 bp,杂合子中一条等位基因目的产物片段为1504 bp,F1和F2的目的产物片段为921 bp。C:单克隆细胞株基因型鉴定一代测序图。红色碱基为PAM位点,红色箭头表示Cas9切割位点;蓝色框内为上游接口处碱基,绿色框内为反转序列,黄色框内为下游接口处碱基。D:野生型克隆、转录起始位点反转的单克隆细胞株eRNA PEARL和Pcdhα基因表达的定量分析。TSS:转录起始位点;WT:野生型;inv:反转。**:P<0.01;***:P<0.001。"
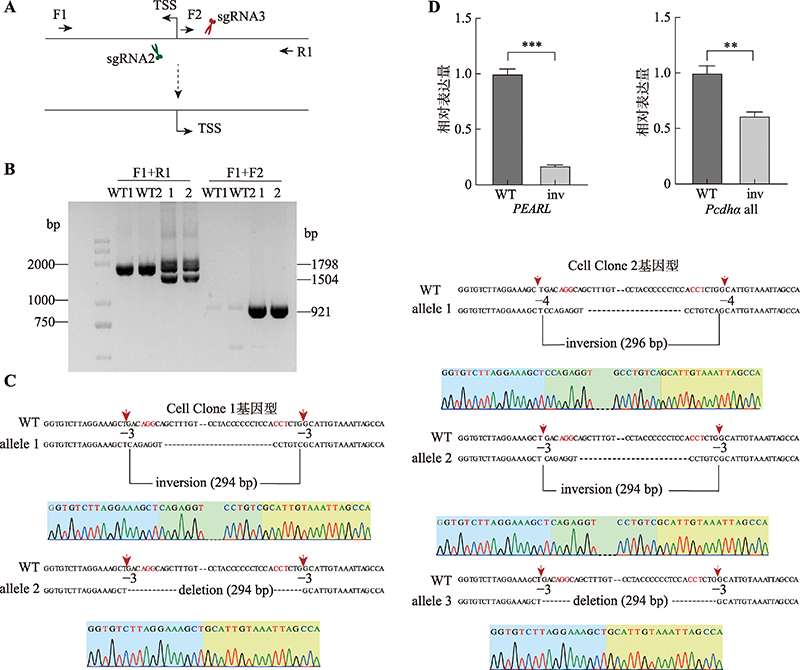
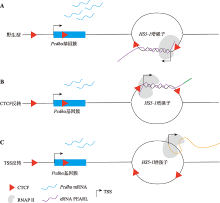
图5
增强子eRNA PEARL调控原钙粘蛋白α基因簇表达的示意图 A:在野生型细胞中,增强子eRNA PEARL (紫色)在HS5-1增强子处形成R环激活Pcdhα基因簇表达。B:使用CRISPR DNA片段编辑系统反转HS5-1增强子及其两边的CTCF位点,不会改变增强子eRNA PEARL转录本的5′端序列(紫色),但会改变eRNA PEARL转录本的3′端序列(绿色),因此可能会改变原本的R环结构,造成Pcdhα基因簇的表达降低。C:反转eRNA PEARL的转录起始位点会完全改变其自身序列(橙色),HS5-1增强子内的R环结构可能遭到破坏,导致Pcdhα基因簇的表达降低。TSS:转录起始位点。"
| [1] | Ong CT, Corces VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat Rev Genet, 2011, 12(4): 283-293. |
| [2] |
Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods, 2013, 10(12): 1213-1218.
doi: 10.1038/nmeth.2688 pmid: 24097267 |
| [3] |
Heinz S, Romanoski CE, Benner C, Glass CK. The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol, 2015, 16(3): 144-154.
doi: 10.1038/nrm3949 |
| [4] |
Banerji J, Rusconi S, Schaffner W. Expression of a β-globin gene is enhanced by remote SV40 DNA sequences. Cell, 1981, 27(<W>2 Pt 1):299-308.
pmid: 6277502 |
| [5] |
Andersson R, Sandelin A. Determinants of enhancer and promoter activities of regulatory elements. Nat Rev Genet, 2020, 21(2): 71-87.
doi: 10.1038/s41576-019-0173-8 pmid: 31605096 |
| [6] |
Furey TS. ChIP-seq and beyond: new and improved methodologies to detect and characterize protein-DNA interactions. Nat Rev Genet, 2012, 13(12): 840-852.
doi: 10.1038/nrg3306 |
| [7] |
Korkmaz G, Lopes R, Ugalde AP, Nevedomskaya E, Han RQ, Myacheva K, Zwart W, Elkon R, Agami R.Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9. Nat Biotechnol, 2016, 34(2): 192-198.
doi: 10.1038/nbt.3450 pmid: 26751173 |
| [8] |
Simeonov DR, Gowen BG, Boontanrart M, Roth TL, Gagnon JD, Mumbach MR, Satpathy AT, Lee YJ, Bray NL, Chan AY, Lituiev DS, Nguyen ML, Gate RE, Subramaniam M, Li ZM, Woo JM, Mitros T, Ray GJ, Curie GL, Naddaf N, Chu JS, Ma H, Boyer E, van Gool F, Huang HL, Liu RZ, Tobin VR, Schumann K, Daly MJ, Farh KK, Ansel KM, Ye CJ, Greenleaf WJ, Anderson MS, Bluestone JA, Chang HY, Corn JE, Marson A. Discovery of stimulation-responsive immune enhancers with CRISPR activation. Nature, 2017, 549(7670): 111-115.
doi: 10.1038/nature23875 |
| [9] |
Klann TS, Black JB, Chellappan M, Safi A, Song LY, Hilton IB, Crawford GE, Reddy TE, Gersbach CA. CRISPR-Cas9 epigenome editing enables high-throughput screening for functional regulatory elements in the human genome. Nat Biotechnol, 2017, 35(6): 561-568.
doi: 10.1038/nbt.3853 |
| [10] |
Calo E, Wysocka J.Modification of enhancer chromatin: what, how, and why? Mol Cell, 2013, 49(5): 825-837.
doi: 10.1016/j.molcel.2013.01.038 |
| [11] |
Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao XB, Schmidl C, Suzuki T, Ntini E, Arner E, Valen E, Li K, Schwarzfischer L, Glatz D, Raithel J, Lilje B, Rapin N, Bagger FO, Jørgensen M, Andersen PR, Bertin N, Rackham O, Burroughs AM, Baillie JK, Ishizu Y, Shimizu Y, Furuhata E, Maeda S, Negishi Y, Mungall CJ, Meehan TF, Lassmann T, Itoh M, Kawaji H, Kondo N, Kawai J, Lennartsson A, Daub CO, Heutink P, Hume DA, Jensen TH, Suzuki H, Hayashizaki Y, Müller F, Forrest ARR, Carninci P, Rehli M, Sandelin A. An atlas of active enhancers across human cell types and tissues. Nature, 2014, 507(7493): 455-461.
doi: 10.1038/nature12787 |
| [12] |
Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, Worley PF, Kreiman G, Greenberg ME. Widespread transcription at neuronal activity-regulated enhancers. Nature, 2010, 465(7295): 182-187.
doi: 10.1038/nature09033 |
| [13] |
Pefanis E, Wang JG, Rothschild G, Lim J, Kazadi D, Sun JB, Federation A, Chao J, Elliott O, Liu ZP, Economides AN, Bradner JE, Rabadan R, Basu U. RNA exosome- regulated long non-coding RNA transcription controls super-enhancer activity. Cell, 2015, 161(4): 774-789.
doi: 10.1016/j.cell.2015.04.034 |
| [14] |
Li WB, Notani D, Rosenfeld MG. Enhancers as non- coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet, 2016, 17(4): 207-223.
doi: 10.1038/nrg.2016.4 |
| [15] |
Li X, Fu XD. Chromatin-associated RNAs as facilitators of functional genomic interactions. Nat Rev Genet, 2019, 20(9): 503-519.
doi: 10.1038/s41576-019-0135-1 |
| [16] |
Sartorelli V, Lauberth SM. Enhancer RNAs are an important regulatory layer of the epigenome. Nat Struct Mol Biol, 2020, 27(6): 521-528.
doi: 10.1038/s41594-020-0446-0 pmid: 32514177 |
| [17] |
Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase Ⅱ. Nat Struct Mol Biol, 2007, 14(2): 103-105.
pmid: 17277804 |
| [18] |
Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature, 2013, 494(7438): 497-501.
doi: 10.1038/nature11884 |
| [19] |
Hsieh CL, Fei T, Chen YW, Li TT, Gao YF, Wang XD, Sun T, Sweeney CJ, Lee GSM, Chen SY, Balk SP, Liu XS, Brown M, Kantoff PW. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc Natl Acad Sci USA, 2014, 111(20): 7319-7324.
doi: 10.1073/pnas.1324151111 |
| [20] |
Li WB, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song XY, Oh S, Kim HS, Glass CK, Rosenfeld MG. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature, 2013, 498(7455): 516-520.
doi: 10.1038/nature12210 |
| [21] |
Xiang JF, Yin QF, Chen T, Zhang Y, Zhang XO, Wu Z, Zhang SF, Wang HB, Ge JH, Lu XH, Yang L, Chen LL. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res, 2014, 24(5): 513-531.
doi: 10.1038/cr.2014.35 |
| [22] |
Bose DA, Donahue G, Reinberg D, Shiekhattar R, Bonasio R, Berger SL. RNA binding to CBP stimulates histone acetylation and transcription. Cell, 2017, 168(1-2): 135-149.e22.
doi: 10.1016/j.cell.2016.12.043 |
| [23] |
Dorighi KM, Swigut T, Henriques T, Bhanu NV, Scruggs BS, Nady N, Still CD, Garcia BA, Adelman K, Wysocka J. Mll3 and Mll4 facilitate enhancer RNA synthesis and transcription from promoters independently of H3K4 monomethylation. Mol Cell, 2017, 66(4): 568-576.
doi: 10.1016/j.molcel.2017.04.018 |
| [24] |
Jia ZL, Wu Q. Clustered protocadherins emerge as novel susceptibility loci for mental disorders. Front Neurosci, 2020, 14: 587819.
doi: 10.3389/fnins.2020.587819 |
| [25] |
Wu Q, Jia ZL. Wiring the brain by clustered protocadherin neural codes. Neurosci Bull, 2021, 37(1): 117-131.
doi: 10.1007/s12264-020-00578-4 |
| [26] |
Wu Q, Maniatis T. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell, 1999, 97(6): 779-790.
pmid: 10380929 |
| [27] |
Tasic B, Nabholz CE, Baldwin KK, Kim Y, Rueckert EH, Ribich SA, Cramer P, Wu Q, Axel R, Maniatis T. Promoter choice determines splice site selection in protocadherin α and γ pre-mRNA splicing. Mol Cell, 2002, 10(1): 21-33.
doi: 10.1016/S1097-2765(02)00578-6 |
| [28] |
Wang XZ, Su H, Bradley A. Molecular mechanisms governing Pcdh-γ gene expression: evidence for a multiple promoter and cis-alternative splicing model. Genes Dev, 2002, 16(15): 1890-1905.
doi: 10.1101/gad.1004802 |
| [29] |
Ribich S, Tasic B, Maniatis T. Identification of long-range regulatory elements in the protocadherin-α gene cluster. Proc Natl Acad Sci USA, 2006, 103(52): 19719-19724.
doi: 10.1073/pnas.0609445104 |
| [30] |
Kehayova P, Monahan K, Chen WS, Maniatis T. Regulatory elements required for the activation and repression of the protocadherin-α gene cluster. Proc Natl Acad Sci USA, 2011, 108(41): 17195-17200.
doi: 10.1073/pnas.1114357108 |
| [31] |
Guo Y, Monahan K, Wu HY, Gertz J, Varley KE, Li W, Myers RM, Maniatis T, Wu Q. CTCF/cohesin-mediated DNA looping is required for protocadherin α promoter choice. Proc Natl Acad Sci USA, 2012, 109(51): 21081-21086.
doi: 10.1073/pnas.1219280110 |
| [32] |
Monahan K, Rudnick ND, Kehayova PD, Pauli F, Newberry KM, Myers RM, Maniatis T. Role of CCCTC binding factor (CTCF) and cohesin in the generation of single-cell diversity of Protocadherin-α gene expression. Proc Natl Acad Sci USA, 2012, 109(23): 9125-9130.
doi: 10.1073/pnas.1205074109 |
| [33] | Zhai YN, Xu Q, Guo Y, Wu Q. Characterization of a cluster of CTCF-binding sites in a protocadherin regulatory region. Hereditas(Beijng), 2016, 38(4): 323-336. |
| 翟亚男, 许泉, 郭亚, 吴强. 原钙粘蛋白基因簇调控区域中成簇的CTCF结合位点分析. 遗传, 2016, 38(4): 323-336. | |
| [34] |
Guo Y, Xu Q, Canzio D, Shou J, Li JH, Gorkin DU, Jung I, Wu HY, Zhai YN, Tang YX, Lu YC, Wu YH, Jia ZL, Li W, Zhang MQ, Ren B, Krainer AR, Maniatis T, Wu Q. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell, 2015, 162(4): 900-910.
doi: 10.1016/j.cell.2015.07.038 pmid: 26276636 |
| [35] |
Lu YJ, Shou J, Jia ZL, Wu YH, Li JH, Guo Y, Wu Q. Genetic evidence for asymmetric blocking of higher-order chromatin structure by CTCF/cohesin. Protein Cell, 2019, 10(12): 914-920.
doi: 10.1007/s13238-019-00656-y |
| [36] |
Jia ZL, Li JW, Ge X, Wu YH, Guo Y, Wu Q. Tandem CTCF sites function as insulators to balance spatial chromatin contacts and topological enhancer-promoter selection. Genome Biol, 2020, 21(1): 75.
doi: 10.1186/s13059-020-01984-7 |
| [37] | Wang N, Jia ZL, Wu Q. RFX5 regulates gene expression of the Pcdhα cluster. Hereditas(Beijng), 2020, 42(8): 760-774. |
| 王娜, 甲芝莲, 吴强. RFX5调控原钙粘蛋白α基因簇的表达. 遗传, 2020, 42(8): 760-774. | |
| [38] |
Tang YX, Jia ZL, Xu HL, Da LT, Wu Q. Mechanism of REST/NRSF regulation of clustered protocadherin α genes. Nucleic Acids Res, 2021, 49(8): 4506-4521.
doi: 10.1093/nar/gkab248 |
| [39] |
Canzio D, Nwakeze CL, Horta A, Rajkumar SM, Coffey EL, Duffy EE, Duffié R, Monahan K, O'Keeffe S, Simon MD, Lomvardas S, Maniatis T. Antisense lncRNA transcription mediates DNA demethylation to drive stochastic protocadherin α promoter choice. Cell, 2019, 177(3): 639-653.e15.
doi: 10.1016/j.cell.2019.03.008 |
| [40] |
Zhou YX, Xu SY, Zhang M, Wu Q. Systematic functional characterization of antisense eRNA of protocadherin alpha composite enhancer. Genes Dev, 2021, 35(19-20): 1383-1394.
doi: 10.1101/gad.348621.121 |
| [41] | Li JH, Shou J, Wu Q.DNA fragment editing of genomes by CRISPR/Cas9. Hereditas(Beijng), 2015, 37(10): 992-1002. |
| 李金环, 寿佳, 吴强. CRISPR/Cas9系统在基因组DNA片段编辑中的应用. 遗传, 2015, 37(10): 992-1002. | |
| [42] |
Shou J, Li JH, Liu YB, Wu Q. Precise and predictable CRISPR chromosomal rearrangements reveal principles of Cas9-mediated nucleotide insertion. Mol Cell, 2018, 71(4): 498-509.e4.
doi: S1097-2765(18)30466-0 pmid: 30033371 |
| [43] | Liu PF, Wu Q.Probing 3D genome by CRISPR/Cas9. Hereditas(Beijng), 2020, 42(1): 18-31. |
| 刘沛峰, 吴强. CRISPR/Cas9基因编辑在三维基因组研究中的应用. 遗传, 2020, 42(1): 18-31. | |
| [44] |
Wu Q, Shou J. Toward precise CRISPR DNA fragment editing and predictable 3D genome engineering. J Mol Cell Biol, 2021, 12(11): 828-856.
doi: 10.1093/jmcb/mjaa060 |
| [45] |
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature, 2012, 489(7414): 57-74.
doi: 10.1038/nature11247 |
| [46] |
Gorkin DU, Barozzi I, Zhao Y, Zhang YX, Huang H, Lee AY, Li B, Chiou J, Wildberg A, Ding B, Zhang B, Wang MC, Strattan JS, Davidson JM, Qiu YJ, Afzal V, Akiyama JA, Plajzer-Frick I, Novak CS, Kato M, Garvin TH, Pham QT, Harrington AN, Mannion BJ, Lee EA, Fukuda- Yuzawa Y, He YP, Preissl S, Chee S, Han JY, Williams BA, Trout D, Amrhein H, Yang HB, Cherry JM, Wang W, Gaulton K, Ecker JR, Shen Y, Dickel DE, Visel A, Pennacchio LA, Ren B. An atlas of dynamic chromatin landscapes in mouse fetal development. Nature, 2020, 583(7818): 744-751.
doi: 10.1038/s41586-020-2093-3 |
| [47] |
García-Muse T, Aguiléra A. R loops: from physiological to pathological roles. Cell, 2019, 179(3): 604-618.
doi: S0092-8674(19)31006-2 pmid: 31607512 |
| [48] |
Niehrs C, Luke B. Regulatory R-loops as facilitators of gene expression and genome stability. Nat Rev Mol Cell Biol, 2020, 21(3): 167-178.
doi: 10.1038/s41580-019-0206-3 |
| [49] |
Suzuki ST. Protocadherins and diversity of the cadherin superfamily. J Cell Sci, 1996, 109(Pt 11): 2609-2611.
doi: 10.1242/jcs.109.11.2609 |
| [50] |
Rahnamoun H, Lee J, Sun ZX, Lu HB, Ramsey KM, Komives EA, Lauberth SM. RNAs interact with BRD4 to promote enhanced chromatin engagement and transcription activation. Nat Struct Mol Biol, 2018, 25(8): 687-697.
doi: 10.1038/s41594-018-0102-0 pmid: 30076409 |
| [51] |
Huang ZQ, Liang N, Goñi S, Damdimopoulos A, Wang C, Ballaire R, Jager J, Niskanen H, Han HY, Jakobsson T, Bracken AP, Aouadi M, Venteclef N, Kaikkonen MU, Fan RR, Treuter E. The corepressors GPS2 and SMRT control enhancer and silencer remodeling via eRNA transcription during inflammatory activation of macrophages. Mol Cell, 2021, 81(5): 953-968.e9.
doi: 10.1016/j.molcel.2020.12.040 |
| [1] | 王舜泽, 江丰, 朱东丽, 杨铁林, 郭燕. Hi-C技术在三维基因组学和疾病致病机理研究中的应用[J]. 遗传, 2023, 45(4): 279-294. |
| [2] | 袁萌, 李辉, 王守志. 大规模平行报告基因测定:一种分析基因表达调控的新技术[J]. 遗传, 2023, 45(10): 859-873. |
| [3] | 赵岩, 王晨鑫, 杨天明, 李春爽, 张丽宏, 杜冬妮, 王若曦, 王静, 魏民, 巴雪青. DNA氧化损伤8-羟鸟嘌呤与肿瘤的发生发展[J]. 遗传, 2022, 44(6): 466-477. |
| [4] | 蒋卓远, 查艳, 石小峰, 张永彪. 神经嵴细胞和神经嵴病及其致病机制的研究进展[J]. 遗传, 2022, 44(2): 117-133. |
| [5] | 毛轲, 孟子秋, 张永彪. 神经嵴发育调控及颅面部遗传基础研究进展[J]. 遗传, 2022, 44(12): 1089-1102. |
| [6] | 朱前彬, 甘志承, 李晓翠, 张英杰, 赵合明, 黄先忠. 小鼠耳芥MAPKKK基因家族全基因组鉴定及进化与表达[J]. 遗传, 2022, 44(11): 1044-1055. |
| [7] | 韩玉婷, 许博文, 李羽童, 卢心怡, 董习之, 邱雨浩, 车沁耘, 朱芮葆, 郑丽, 李孝宸, 司绪, 倪建泉. 模式动物果蝇的基因调控前沿技术[J]. 遗传, 2022, 44(1): 3-14. |
| [8] | 周聪, 周强伟, 成盛, 李国亮. CTCF在介导三维基因组形成及调控基因表达中的研究进展[J]. 遗传, 2021, 43(9): 816-821. |
| [9] | 林红燕, 王煊, 何聪, 周紫玲, 杨旻恺, 文钟灵, 韩洪苇, 陆桂华, 戚金亮, 杨永华. 中药植物紫草天然产物的生物合成及其功能研究进展[J]. 遗传, 2021, 43(5): 459-472. |
| [10] | 徐海冬, 宁博林, 牟芳, 李辉, 王宁. 选择性多聚腺苷酸化的生物学效应及其调控机制研究进展[J]. 遗传, 2021, 43(1): 4-15. |
| [11] | 王涛涛, 杨勇, 魏唯, 林辰涛, 马留银. 互花米草NAC转录因子家族的鉴定与表达分析[J]. 遗传, 2020, 42(2): 194-211. |
| [12] | 陈会友, 张建敏, 李柏森, 邓永琳, 张龚炜. 犏牛雄性不育的减数分裂基因表达与表观遗传调控研究进展[J]. 遗传, 2020, 42(11): 1081-1092. |
| [13] | 高晓萌, 张治华. 生物大分子“液-液相分离”调控染色质三维空间结构和功能[J]. 遗传, 2020, 42(1): 45-56. |
| [14] | 禹奇超,宋彬,邹轩轩,王岭,刘德权,李波,马昆. 乳腺癌癌旁组织特异性表达基因分析[J]. 遗传, 2019, 41(7): 625-633. |
| [15] | 邢万金. 乳糖操纵子模型的建立与教学中若干问题的解析[J]. 遗传, 2019, 41(6): 548-563. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||

www.chinagene.cn
备案号:京ICP备09063187号-4
总访问:,今日访问:,当前在线:
