遗传 ›› 2022, Vol. 44 ›› Issue (9): 783-797.doi: 10.16288/j.yczz.22-201
收稿日期:2022-06-16
修回日期:2022-07-20
出版日期:2022-09-20
发布日期:2022-08-01
作者简介:陈秀丽,硕士研究生,专业方向:生物学。E-mail: 基金资助:
Xiuli Chen( ), Haiyan Huang(
), Haiyan Huang( ), Qiang Wu(
), Qiang Wu( )
)
Received:2022-06-16
Revised:2022-07-20
Published:2022-09-20
Online:2022-08-01
Supported by:摘要:
人类β-地中海贫血的发病机制与β-样珠蛋白基因异常表达息息相关。人类β-样珠蛋白基因以5′-ε-Gγ-Aγ-δ-β-3′的顺序排列于β-珠蛋白基因座,受5′LCR (locus control region)中5个超敏位点(hypersensitive site, HS)5′HS5~5′HS1和3′HS1调控。其中5′HS2是最重要的增强子,能产生增强子RNA (enhancer RNA)并调控ε-globin、γ-globin和β-globin的表达。为了进一步探究K562细胞中增强子5′HS2的功能,本研究首先通过染色质构象捕获技术在人慢性髓原白血病K562细胞中探测到5′HS2介导的染色质相互作用集中在以包含CTCF (CCCTC-binding factor)位点的3′HS1和5′HS5为边界的拓扑结构域中,5′HS2在三维空间上与HBE1、HBG2和HBG1启动子区域相互靠近。其次运用CRISPR DNA片段编辑技术在K562细胞系中删除了增强子5′HS2。最后通过RNA-seq和CUT&Tag (cleavage under target & tagmentation)实验分析两个5′HS2删除的单克隆细胞系的转录组和染色质H3K27ac组蛋白修饰,发现91个基因表达显著下调而且其启动子区的H3K27ac修饰程度显著降低。这些基因主要聚类于氧气运输、免疫应答、细胞粘附、抗氧化和维持血栓形成等与红细胞功能相关的生物过程中,表明K562细胞系中增强子5′HS2对红细胞功能相关基因的转录产生了广泛影响。
陈秀丽, 黄海燕, 吴强. 靶向敲除β-珠蛋白基因座控制区增强子HS2对K562细胞转录组的影响[J]. 遗传, 2022, 44(9): 783-797.
Xiuli Chen, Haiyan Huang, Qiang Wu. Targeted deletion of 5′HS2 enhancer of β-globin locus control region in K562 cells[J]. Hereditas(Beijing), 2022, 44(9): 783-797.
表1
引物序列"
| 引物类型 | 引物名称 | 引物序列(5′→3′) |
|---|---|---|
| sgRNA | hHB_EHS2-sg1F | caccTCTGCCGCTTCTAGGTATAG |
| hHB_EHS2-sg1R | aaacCTATACCTAGAAGCGGCAGA | |
| hHB_EHS2-sg2F | caccCTTAGTCTTAGAGGCCCCAC | |
| hHB_EHS2-sg2R | aaacGTGGGGCCTCTAAGACTAAG | |
| PCR | universal-gRNA_R | GCACCGACTCGGTGCCACTT |
| hHB_EHS2- genotyping-F1 | GTAGTCCTTCACAGTTACCCACACA | |
| hHB_EHS2-genotyping-R1 | CATTAGTGACCTCCCATAGTCCAAG | |
| hHB_EHS2-genotyping-F2 | CCTGAGCTCCAAGCAGTCCAC | |
| hHB_EHS2-genotyping-R2 | CATTTGGCCCCTCCTAATCTCTC | |
| biotin修饰引物 | biotin-hs2[ | 5′biotin-CAGTTACCCACACAGGTGAACCC |
| P5引物 | qhr-hs2-F1[ | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTGGAATGTTTCTTTCCTCTCAGGATC |
| P7引物 | wly-P2 | CAAGCAGAAGACGGCATACGAGATTTCATGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT |
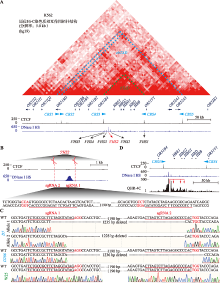
图1
K562细胞中CRISPR删除β-珠蛋白LCR中的关键增强子元件5′HS2 A:K562细胞系中β-珠蛋白基因座相关的原位Hi-C (high-through chromosome conformation capture)染色质拓扑结构域、CTCF (CCCTC-binding factor) ChIP-seq结合峰分布图和指示染色质开放程度的DNase I超敏位点(hypersensitive site, HS)分布。β-珠蛋白基因座包含5个基因,HBE1、HBG2、HBG1、HBD和HBB;bTAD (β-globin TAD)和ebTAD (extended β-globin TAD)的范围分别用绿色和蓝色虚线标出;黑色或红色箭头标注调控元件3′HS1、5′HS1、5′HS2、5′HS3、5′HS4和5′HS5,蓝色三角箭头标注CBS (CTCF binding site)的方向。B:增强子5′HS2删除实验中两个sgRNA的靶向序列及位置。红色字母突显PAM (protospacer adjacent motif)序列,红色横线指示靶向序列。C:5′HS2删除克隆D7、D286和对照未被编辑的克隆(野生型克隆) W25的基因型鉴定结果。克隆D7中检测到两种等位基因Allele 1和Allele 2,分别删除了包含5′HS2的1235 bp和1228 bp片段;克隆D286中只检测到一种等位基因,删除了包含5′HS2的1236 bp片段。D:以5′HS2邻近序列为锚定点的染色质构象捕获结果。参考DNase I超敏位点分布可确定5′HS2位置;红色箭头标出了与5′HS2相互作用的基因;参考CTCF ChIP-seq结合峰分布图标出了CBS3和CBS4。"
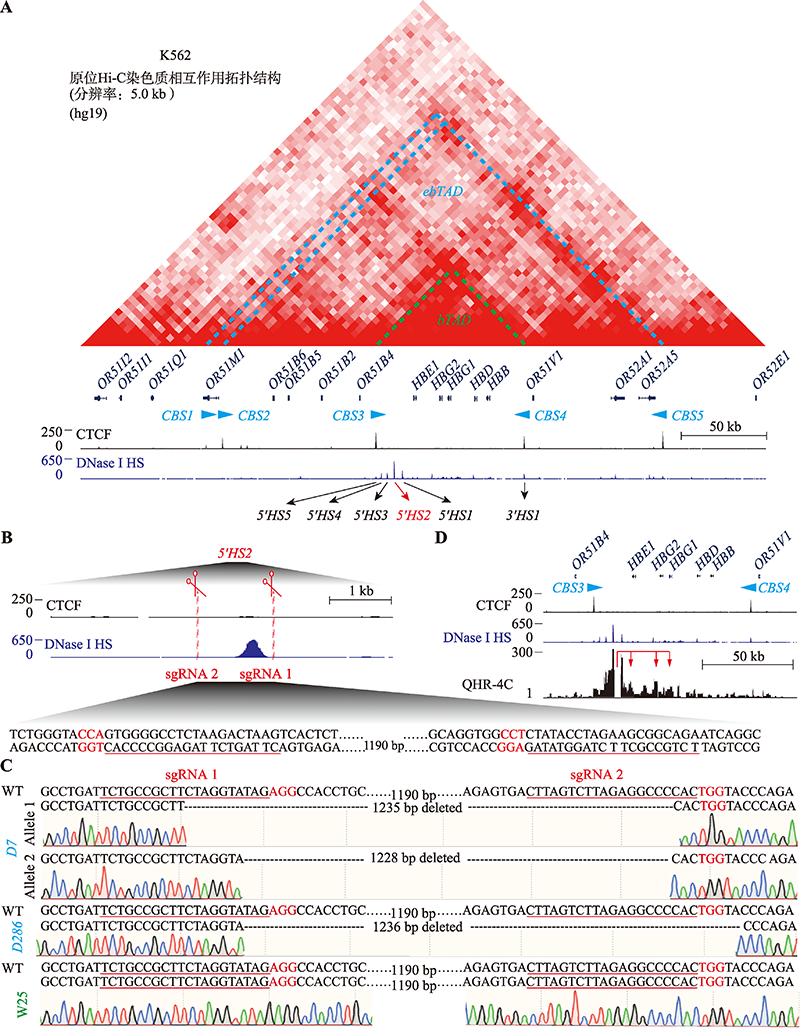
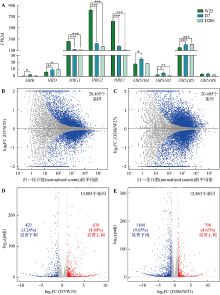
图2
RNA-seq检测靶向删除增强子5′HS2对K562细胞转录组的影响 A:RNA-seq检测增强子5′HS2删除后ebTAD范围内基因的转录水平变化。W25、D7、D286分别代表野生型和两个5′HS2删除型单克隆细胞株;每个样本进行3个生物学重复;FPKM:fragments per kilobase per million mapped reads;*:P < 0.05;**:P < 0.01;***:P < 0.001。B,C:克隆D7(B)和D286(C)相较对照克隆W25的差异表达基因MA图。蓝点表示差异基因,灰点表示其他基因。D,E:克隆D7(D)和D286(E)相较克隆W25的差异表达基因火山图。以log2 FC(log2 fold change)为横坐标,以-log10 (padj)(adjusted P value)为纵坐标;红点表示上调基因,蓝点表示下调基因,灰点表示其余基因;克隆D7差异表达基因分析中保留了13,005个基因,其中有630(4.84%)个基因上调和421(3.24%)个基因下调;克隆D286差异表达基因分析中保留了12,863个基因,其中有586(4.63%)个基因上调和1164(9.05%)个基因下调。"
| [1] | Hill RJ, Konigsberg W, Guidotti G, Craig LC . The structure of human hemoglobin. I. The separation of the alpha and beta chains and their amino acid composition. J Biol Chem, 1962,237:1549-1554. |
| [2] | Rahbar S, Beale D, Isaacs WA, Lehmann H . Abnormal haemoglobins in Iran. Observation of a new variant— haemoglobin J Iran (alpha-2-beta-2 77 His→Asp). Br Med J, 1967,1(5541):674-677. |
| [3] | Anson ML, Mirsky AE . On hemochromogen. J Gen Physiol, 1928,12(2):273-288. |
| [4] | Origa R . β-Thalassemia. Genet Med, 2017,19(6):609-619. |
| [5] | Maquat LE, Kinniburgh AJ, Rachmilewitz EA, Ross J . Unstable beta-globin mRNA in mRNA-deficient beta o thalassemia. Cell, 1981,27(3 Pt 2):543-553. |
| [6] | Ingram VM . Gene mutations in human haemoglobin: the chemical difference between normal and sickle cell haemoglobin. Nature, 1957,180(4581):326-328. |
| [7] | Grosveld F, Antoniou M, Berry M, De Boer E, Dillon N, Ellis J, Fraser P, Hanscombe O, Hurst J, Imam A, Lindenbaum M, Philipsen S, Pruzina S, Strouboulis J, Raguz-Bolognesi S, Talbot D . The regulation of human globin gene switching. Philos Trans R Soc Lond B Biol Sci, 1993,339(1288):183-191. |
| [8] | Guo Y, Xu Q, Canzio D, Shou J, Li JH, Gorkin DU, Jung I, Wu HY, Zhai YN, Tang YX, Lu YC, Wu YH, Jia ZL, Li W, Zhang MQ, Ren B, Krainer AR, Maniatis T, Wu Q . CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell, 2015,162(4):900-910. |
| [9] | Himadewi P, Wang XQD, Feng F, Gore H, Liu YS, Yu L, Kurita R, Nakamura Y, Pfeifer GP, Liu J, Zhang XT . 3′HS1 CTCF binding site in human β-globin locus regulates fetal hemoglobin expression. eLife, 2021,10:e70557. |
| [10] | Wai AWK, Gillemans N, Raguz-Bolognesi S, Pruzina S, Zafarana G, Meijer D, Philipsen S, Grosveld F . HS5 of the human beta-globin locus control region: a developmental stage-specific border in erythroid cells. EMBO J, 2003,22(17):4489-4500. |
| [11] | Pasceri P, Pannell D, Wu X, Ellis J . Full activity from human beta-globin locus control region transgenes requires 5′HS1, distal beta-globin promoter, and 3′ beta-globin sequences. Blood, 1998,92(2):653-663. |
| [12] | Fedosyuk H, Peterson KR . Deletion of the human beta-globin LCR 5′HS4 or 5′HS1 differentially affects beta-like globin gene expression in beta-YAC transgenic mice. Blood Cells Mol Dis, 2007,39(1):44-55. |
| [13] | Peterson KR, Clegg CH, Navas PA, Norton EJ, Kimbrough TG, Stamatoyannopoulos G . Effect of deletion of 5'HS3 or 5′HS2 of the human beta-globin locus control region on the developmental regulation of globin gene expression in beta-globin locus yeast artificial chromosome transgenic mice. Proc Natl Acad Sci USA, 1996,93(13):6605-6609. |
| [14] | Navas PA, Peterson KR, Li Q, Skarpidi E, Rohde A, Shaw SE, Clegg CH, Asano H, Stamatoyannopoulos G . Developmental specificity of the interaction between the locus control region and embryonic or fetal globin genes in transgenic mice with an HS3 core deletion. Mol Cell Biol, 1998,18(7):4188-4196. |
| [15] | Kim S, Kim YW, Shim SH, Kim CG, Kim A . Chromatin structure of the LCR in the human β-globin locus transcribing the adult δ- and β-globin genes. Int J Biochem Cell Biol, 2012,44(3):505-513. |
| [16] | Navas PA, Peterson KR, Li Q, McArthur M, Stamatoyannopoulos G. The 5′HS4 core element of the human beta-globin locus control region is required for high-level globin gene expression in definitive but not in primitive erythropoiesis. J Mol Biol, 2001,312(1):17-26. |
| [17] | Zhou YX, Xu SY, Zhang M, Wu Q . Systematic functional characterization of antisense eRNA of protocadherin α composite enhancer. Genes Dev, 2021,35(19-20):1383-1394. |
| [18] | Lee JH, Wang RY, Xiong F, Krakowiak J, Liao ZA, Nguyen PT, Moroz-Omori EV, Shao JF, Zhu XY, Bolt MJ, Wu HY, Singh PK, Bi MJ, Shi CJ, Jamal N, Li GJ, Mistry R, Jung SY, Tsai KL, Ferreon JC, Stossi F, Caflisch A, Liu ZJ, Mancini MA, Li WB. Enhancer RNA m6A methylation facilitates transcriptional condensate formation and gene activation. Mol Cell, 2021, 81(16): 3368-3385.e9. |
| [19] | Gurumurthy A, Yu DT, Stees JR, Chamales P, Gavrilova E, Wassel P, Li L, Stribling D, Chen JY, Brackett M, Ishov AM, Xie MY, Bungert J . Super-enhancer mediated regulation of adult β-globin gene expression: the role of eRNA and Integrator. Nucleic Acids Res, 2021,49(3):1383-1396. |
| [20] | Cho Y, Song SH, Lee JJ, Choi N, Kim CG, Dean A, Kim A . The role of transcriptional activator GATA-1 at human beta-globin HS2. Nucleic Acids Res, 2008,36(14):4521-4528. |
| [21] | Kang YJ, Kim YW, Kang J, Yun WJ, Kim A . Erythroid specific activator GATA-1-dependent interactions between CTCF sites around the β-globin locus. Biochim Biophys Acta Gene Regul Mech, 2017,1860(4):416-426. |
| [22] | Kim J, Kang J, Kim YW, Kim A . The human β-globin enhancer LCR HS2 plays a role in forming a TAD by activating chromatin structure at neighboring CTCF sites. FASEB J, 2021,35(6):e21669. |
| [23] | Jia ZL, Li JW, Ge X, Wu YH, Guo Y, Wu Q . Tandem CTCF sites function as insulators to balance spatial chromatin contacts and topological enhancer-promoter selection. Genome Biol, 2020,21(1):75. |
| [24] | Martin M . Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J, 2011,17(1):10-12. |
| [25] | Kim D, Langmead B, Salzberg SL . HISAT: a fast spliced aligner with low memory requirements. Nat Methods, 2015,12(4):357-360. |
| [26] | Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol, 2010,28(5):511-515. |
| [27] | Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL . StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol, 2015,33(3):290-295. |
| [28] | Love MI, Huber W, Anders S . Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol, 2014,15(12):550. |
| [29] | Wu TZ, Hu EQ, Xu SB, Chen MJ, Guo PF, Dai ZH, Feng TZ, Zhou L, Tang WL, Zhan L, Fu XC, Liu SS, Bo XC, Yu GC. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (Camb), 2021,2(3):100141. |
| [30] | Chen L, Zhang YH, Wang SP, Zhang YH, Huang T, Cai YD . Prediction and analysis of essential genes using the enrichments of gene ontology and KEGG pathways. PLoS One, 2017,12(9):e0184129. |
| [31] | Langmead B, Salzberg SL . Fast gapped-read alignment with Bowtie 2. Nat Methods, 2012,9(4):357-359. |
| [32] | Grytten I, Rand KD, Nederbragt AJ, Storvik GO, Glad IK, Sandve GK . Graph Peak Caller: calling ChIP-seq peaks on graph-based reference genomes. PLoS Comput Biol, 2019,15(2):e1006731. |
| [33] | Guo Y, Monahan K, Wu HY, Gertz J, Varley KE, Li W, Myers RM, Maniatis T, Wu Q . CTCF/cohesin-mediated DNA looping is required for protocadherin α promoter choice. Proc Natl Acad Sci USA, 2012,109(51):21081-21086. |
| [34] | Ziebarth JD, Bhattacharya A, Cui Y . CTCFBSDB 2.0: a database for CTCF-binding sites and genome organization. Nucleic Acids Res, 2013,41(Database issue):D188-D194. |
| [35] | Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, Aiden EL . A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell, 2014,159(7):1665-1680. |
| [36] | Wu Q, Shou J . Toward precise CRISPR DNA fragment editing and predictable 3D genome engineering. J Mol Cell Biol, 2021,12(11):828-856. |
| [37] | Hawkins RD, Hon GC, Yang CH, Antosiewicz-Bourget JE, Lee LK, Ngo QM, Klugman S, Ching KA, Edsall LE, Ye Z, Kuan S, Yu PZ, Liu H, Zhang XM, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Ren B . Dynamic chromatin states in human ES cells reveal potential regulatory sequences and genes involved in pluripotency. Cell Res, 2011,21(10):1393-1409. |
| [38] | Wang ZB, Zang CZ, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui KR, Roh TY, Peng WQ, Zhang MQ, Zhao KJ . Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet, 2008,40(7):897-903. |
| [39] | Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G . Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet, 2000,25(1):25-29. |
| [40] | Klein E, Ben-Bassat H, Neumann H, Ralph P, Zeuthen J, Polliack A, Vánky F . Properties of the K562 cell line, derived from a patient with chronic myeloid leukemia. Int J Cancer, 1976,18(4):421-431. |
| [41] | Andersson LC, Nilsson K, Gahmberg CG . K562--a human erythroleukemic cell line. Int J Cancer, 1979,23(2):143-147. |
| [42] | Cheng ML, Ho HY, Tseng HC, Lee CH, Shih LY, Chiu DTY . Antioxidant deficit and enhanced susceptibility to oxidative damage in individuals with different forms of alpha-thalassaemia. Br J Haematol, 2005,128(1):119-127. |
| [43] | Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, Eskiw CH, Luo YQ, Wei CL, Ruan YJ, Bieker JJ, Fraser P . Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet, 2010,42(1):53-61. |
| [1] | 安梦婷, 郭冠麟, 吴杰, 孙文靖, 贾学渊. 基于生物信息学分析胃癌双微体中增强子的调控机制[J]. 遗传, 2025, 47(5): 558-572. |
| [2] | 李轲, 周晓蓉, 朱东丽, 陈晓峰, 郭燕. 系统性红斑狼疮易感区域FAM167A-BLK遗传变异的调控机制研究[J]. 遗传, 2025, 47(11): 1244-1255. |
| [3] | 杨敏, 林思远, 杨长淇, 陈瑶生, 何祖勇. SOX9及其增强子在哺乳动物性别决定中的研究进展[J]. 遗传, 2024, 46(9): 677-689. |
| [4] | 王纪龙, 李青, 战廷正. 自转录活性调节区测序技术在增强子发现研究中的应用[J]. 遗传, 2024, 46(8): 589-602. |
| [5] | 王舜泽, 江丰, 朱东丽, 杨铁林, 郭燕. Hi-C技术在三维基因组学和疾病致病机理研究中的应用[J]. 遗传, 2023, 45(4): 279-294. |
| [6] | 徐思远, 寿佳, 吴强. HS5-1增强子eRNA PEARL对原钙粘蛋白α基因簇的表达调控[J]. 遗传, 2022, 44(8): 695-764. |
| [7] | 漆思晗, 王棨临, 张俊有, 刘倩, 李春燕. 增强子调控癌症发生发展的机制研究[J]. 遗传, 2022, 44(4): 275-288. |
| [8] | 万星琦, 魏婉珍, 郭胜良, 崔一笑, 景雪莹, 黄露杰, 马捷. BMP2基因远程调控元件的功能分析[J]. 遗传, 2022, 44(12): 1141-1147. |
| [9] | 周聪, 周强伟, 成盛, 李国亮. CTCF在介导三维基因组形成及调控基因表达中的研究进展[J]. 遗传, 2021, 43(9): 816-821. |
| [10] | 王玲, 李金环, 黄海燕, 吴强. 串联反向CTCF位点的系列删除揭示增强子调控HOXD基因簇表达的平衡[J]. 遗传, 2021, 43(8): 775-791. |
| [11] | 罗鑫, 宿兵. 三维基因组分析点亮人类大脑进化之谜[J]. 遗传, 2021, 43(2): 105-107. |
| [12] | 刘倩, 李春燕. 增强子的鉴定及其在肿瘤研究中的应用[J]. 遗传, 2020, 42(9): 817-831. |
| [13] | 秦中勇, 石晓, 曹平平, 褚鹰, 管蔚, 杨楠, 程禾, 孙玉洁. 细胞凋亡反应中NOXA基因启动子发挥增强子功能调节BCL2基因表达[J]. 遗传, 2020, 42(11): 1110-1121. |
| [14] | 杨科, 薛征, 吕湘. 细胞终末分化过程中三维基因组结构与功能调控的分子机制[J]. 遗传, 2020, 42(1): 32-44. |
| [15] | 张雨, 方玉达. Cohesin结构及功能研究进展[J]. 遗传, 2020, 42(1): 57-72. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||

www.chinagene.cn
备案号:京ICP备09063187号-4
总访问:,今日访问:,当前在线:
