遗传 ›› 2023, Vol. 45 ›› Issue (8): 632-642.doi: 10.16288/j.yczz.23-045
收稿日期:2023-03-01
修回日期:2023-06-21
出版日期:2023-08-20
发布日期:2023-07-04
通讯作者:
沈彬
E-mail:songruijia@stu.njmu.edu.cn;hanlu@stu.njmu.edu.cn;binshen@njmu.edu.cn.
作者简介:宋睿嘉,在读本科生,专业方向:临床医学。E-mail: 基金资助:
Ruijia Song( ), Lu Han(
), Lu Han( ), Haifeng Sun, Bin Shen(
), Haifeng Sun, Bin Shen( )
)
Received:2023-03-01
Revised:2023-06-21
Online:2023-08-20
Published:2023-07-04
Contact:
Bin Shen
E-mail:songruijia@stu.njmu.edu.cn;hanlu@stu.njmu.edu.cn;binshen@njmu.edu.cn.
Supported by:摘要:
线粒体作为真核高等生物的能量工厂,通过有氧呼吸的方式为各项生命活动提供能量(ATP)。线粒体拥有一套独立于细胞核的基因组——线粒体DNA(mitochondrial DNA,mtDNA),编码37个基因,其突变会导致线粒体疾病,目前已在人mtDNA中鉴定出了超过100种致病突变位点,总发病率约为1/5000。近年来,基于CRISPR的碱基编辑技术已经实现了对核基因组的精确编辑,然而由于CRISPR系统中的引导RNA难以通过线粒体的双层膜结构,在mtDNA上实现精确的碱基编辑仍具有较大的挑战性。2020年,美国哈佛大学David R. Liu实验室报道了一种伯克霍尔德氏菌来源的双链DNA脱氨酶DddA,将其与可编程的转录激活样效应因子(transcription activator-like effector,TALE)和尿嘧啶糖苷酶抑制剂(uracil glycosylase inhibitor,UGI)融合组装成为DdCBEs(DddA来源的胞嘧啶碱基编辑器),首次在mtDNA上实现了特异高效的C·G到T·A的转换。本文对近几年基于DddA的线粒体碱基编辑技术的发展进行综述,并对其未来应用前景进行展望,以期为相关领域的科研人员进一步了解、使用及优化线粒体碱基编辑技术提供参考。
宋睿嘉, 韩露, 孙海峰, 沈彬. 线粒体DNA碱基编辑技术研究进展[J]. 遗传, 2023, 45(8): 632-642.
Ruijia Song, Lu Han, Haifeng Sun, Bin Shen. Advances in mitochondrial DNA base editing technology[J]. Hereditas(Beijing), 2023, 45(8): 632-642.
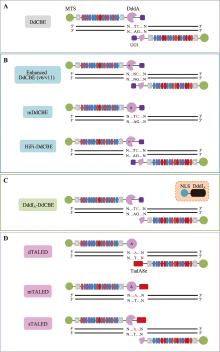
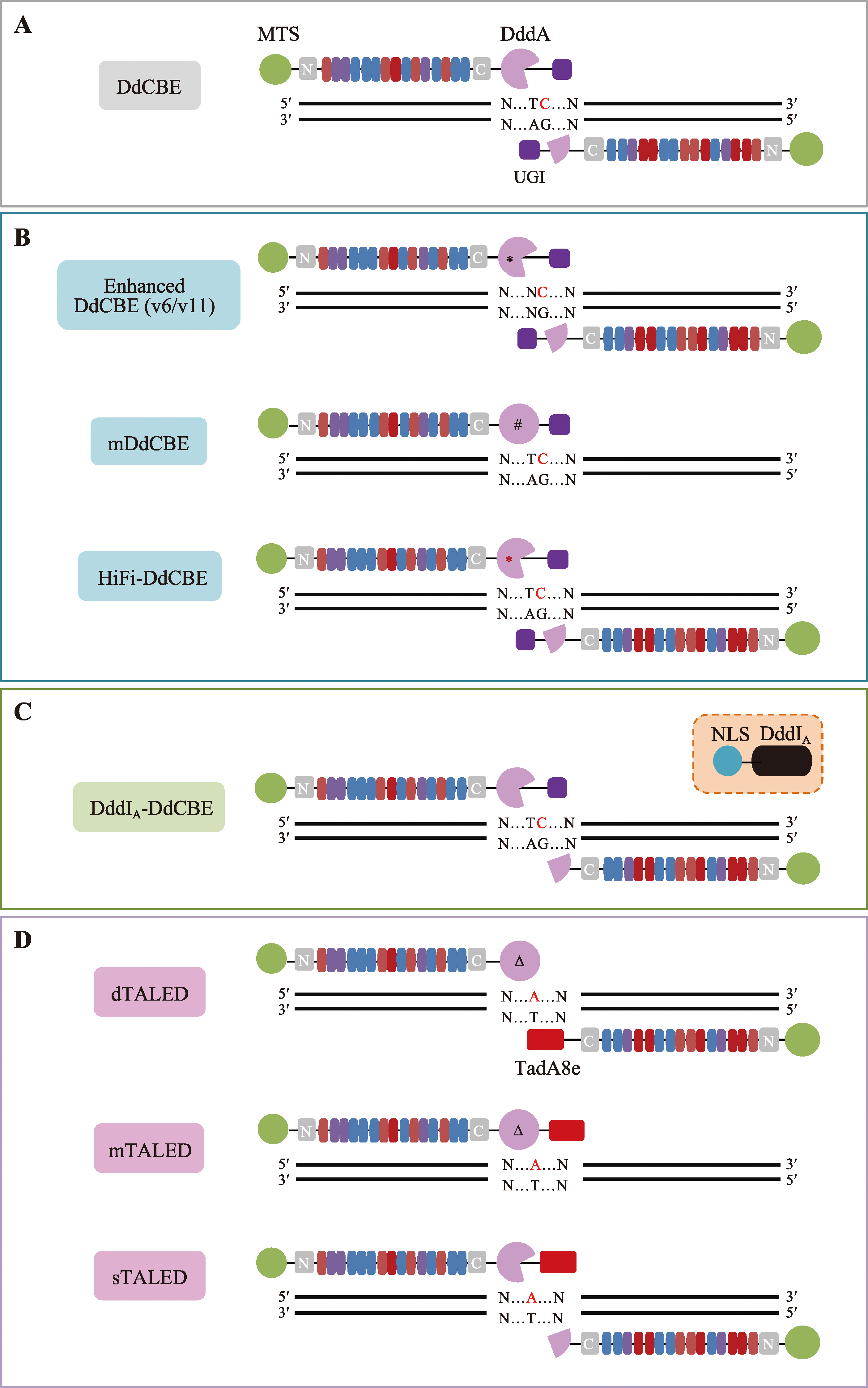
图2
基于DddA开发的线粒体碱基编辑器 A: 原始版本的DdCBEs主要实现TC基序中的C到T的编辑;B:通过对DdCBEs中的DddAtox进行工程化改造得到的不同突变体DdCBEs版本,分别是对TC编辑活性有提升的v6版本(DdCBE-DddA6)、对non-TC基序具有活性的v11版本(DdCBE-DddA11)、携带突变DddAtox的单TALE臂版本(mDdCBE),以及高保真版本(HiFi-DdCBE);C:DddIA-DdCBE通过同时表达核定位的DddIA蛋白,最大程度降低了DdCBE在核内的脱靶;D:通过与腺嘌呤脱氨酶TadA8e结合,构成三种版本的TALEDs,可以实现mtDNA上的A·T到G·C的编辑。黑色星号(*):Mok等[59]通过定向进化得到的增强版DddAtox;黑色井号(#):Mok等[44]工程化改造的没有明显毒性的DddAtox;红色星号(*):Lee等[56]通过界面氨基酸突变得到的高保真型DddAtox;黑色三角号(Δ):催化活性失活的DddAtox。"
| [1] | Jia ZW. Mitochondria and pluripotent stem cells function. Hereditas(Beijing), 2016, 38(7): 603-611. |
| 贾振伟. 线粒体与多潜能干细胞功能. 遗传, 2016, 38(7): 603-611. | |
| [2] |
Tuppen HAL, Blakely EL, Turnbull DM, Taylor RW. Mitochondrial DNA mutations and human disease. Biochim Biophys Acta, 2010, 1797(2): 113-128.
doi: 10.1016/j.bbabio.2009.09.005 pmid: 19761752 |
| [3] |
Neupert W. SnapShot: mitochondrial architecture. Cell, 2012, 149(3): 722-722.e1.
doi: 10.1016/j.cell.2012.04.010 pmid: 22541440 |
| [4] | Schatz G. Mitochondrial DNA. In:Encyclopedia of biological chemistry. Elsevier, 2013, 132-134. |
| [5] | Gray MW. Mitochondrial DNA. In:Brenner’s encyclopedia of genetics. Elsevier, 2013, 436-438. |
| [6] |
Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG. Sequence and organization of the human mitochondrial genome. Nature, 1981, 290(5806): 457-465.
doi: 10.1038/290457a0 |
| [7] |
Ziada AS, Smith MSR, Côté HCF. Updating the free radical theory of aging. Front Cell Dev Biol, 2020, 8: 575645.
doi: 10.3389/fcell.2020.575645 |
| [8] |
Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Genet, 2005, 6(5): 389-402.
doi: 10.1038/nrg1606 pmid: 15861210 |
| [9] |
Biesalski HK. Free radical theory of aging. Curr Opin Clin Nutr Metab Care, 2002, 5(1): 5-10.
doi: 10.1097/00075197-200201000-00002 |
| [10] |
Filograna R, Mennuni M, Alsina D, Larsson NG. Mitochondrial DNA copy number in human disease: the more the better? FEBS Lett, 2021, 595(8): 976-1002.
doi: 10.1002/1873-3468.14021 pmid: 33314045 |
| [11] |
Silva-Pinheiro P, Minczuk M. The potential of mitochondrial genome engineering. Nat Rev Genet, 2022, 23(4): 199-214.
doi: 10.1038/s41576-021-00432-x |
| [12] |
Gammage PA, Moraes CT, Minczuk M. Mitochondrial genome engineering: the revolution may not be CRISPR-Ized. Trends Genet, 2018, 34(2): 101-110.
doi: S0168-9525(17)30191-9 pmid: 29179920 |
| [13] | Pinheiro P, Gammage PA, Minczuk M. Mitochondrially targeted zinc finger nucleases. In: The human mitochondrial genome. Elsevier, 2020, 499-514. |
| [14] |
Gammage PA, Rorbach J, Vincent AI, Rebar EJ, Minczuk M. Mitochondrially targeted ZFNs for selective degradation of pathogenic mitochondrial genomes bearing large- scale deletions or point mutations. EMBO Mol Med, 2014, 6(4): 458-466.
doi: 10.1002/emmm.201303672 pmid: 24567072 |
| [15] |
Gammage PA, Viscomi C, Simard ML, Costa ASH, Gaude E, Powell CA, Van Haute L, McCann BJ, Rebelo-Guiomar P, Cerutti R, Zhang L, Rebar EJ, Zeviani M, Frezza C, Stewart JB, Minczuk M. Genome editing in mitochondria corrects a pathogenic mtDNA mutation in vivo. Nat Med, 2018, 24(11): 1691-1695.
doi: 10.1038/s41591-018-0165-9 pmid: 30250142 |
| [16] |
Gammage PA, Gaude E, Van Haute L, Rebelo-Guiomar P, Jackson CB, Rorbach J, Pekalski ML, Robinson AJ, Charpentier M, Concordet JP, Frezza C, Minczuk M. Near-complete elimination of mutant mtDNA by iterative or dynamic dose-controlled treatment with mtZFNs. Nucleic Acids Res, 2016, 44(16): 7804-7816.
doi: 10.1093/nar/gkw676 pmid: 27466392 |
| [17] | McCann BJ, Cox A, Gammage PA, Stewart JB, Zernicka-Goetz M, Minczuk M. Delivery of mtZFNs into early mouse embryos. In: Liu J (ed). Zinc Finger Proteins, vol. 1867. New York, NY: Springer New York, 2018, 215-228. |
| [18] | Minczuk M. Engineered zinc finger proteins for manipulation of the human mitochondrial genome. In: Mackay JP, Segal DJ eds. Engineered Zinc Finger Proteins vol. 649. Totowa, NJ: Humana Press, 2010, 257-270. |
| [19] | Gammage PA, Van Haute L, Minczuk M. Engineered mtZFNs for manipulation of human mitochondrial DNA heteroplasmy. In: McKenzie M ed. MitochondrialDNA, vol. 1351. New York, NY: Springer New York, 2016, 145-162. |
| [20] |
Reddy P, Ocampo A, Suzuki K, Luo JP, Bacman SR, Williams SL, Sugawara A, Okamura D, Tsunekawa Y, Wu J, Lam D, Xiong X, Montserrat N, Esteban CR, Liu GH, Sancho-Martinez I, Manau D, Civico S, Cardellach F, del Mar O’Callaghan M, Campistol J, Zhao HM, Campistol JM, Moraes CT, Izpisua Belmonte JC. Selective elimination of mitochondrial mutations in the germline by genome editing. Cell, 2015, 161(3): 459-469.
doi: S0092-8674(15)00371-2 pmid: 25910206 |
| [21] |
Hashimoto M, Bacman SR, Peralta S, Falk MJ, Chomyn A, Chan DC, Williams SL, Moraes CT. MitoTALEN: A general approach to reduce mutant mtDNA loads and restore oxidative phosphorylation function in mitochondrial diseases. Mol Ther, 2015, 23(10): 1592-1599.
doi: 10.1038/mt.2015.126 pmid: 26159306 |
| [22] | Bacman SR, Gammage PA, Minczuk M, Moraes CT. Manipulation of mitochondrial genes and mtDNA heteroplasmy. In: Methods in cell biology vol. 155. Elsevier, 2020, 441-487. |
| [23] |
Bacman SR, Kauppila JHK, Pereira CV, Nissanka N, Miranda M, Pinto M, Williams SL, Larsson NG, Stewart JB, Moraes CT. MitoTALEN reduces mutant mtDNA load and restores TRNAAla levels in a mouse model of heteroplasmic mtDNA mutation. Nat Med, 2018, 24(11): 1696-1700.
doi: 10.1038/s41591-018-0166-8 |
| [24] |
Bacman SR, Williams SL, Pinto M, Peralta S, Moraes CT. Specific elimination of mutant mitochondrial genomes in patient-derived cells by mitoTALENs. Nat Med, 2013, 19(9): 1111-1113.
doi: 10.1038/nm.3261 pmid: 23913125 |
| [25] |
Mikhailov N, Hämäläinen RH. Modulating mitochondrial DNA heteroplasmy with mitochondrially targeted endonucleases. Ann Biomed Eng, 2022, doi: 10.1007/s10439-022-03051-7.
doi: 10.1007/s10439-022-03051-7 |
| [26] |
Rees HA, Liu DR. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat Rev Genet, 2018, 19(12): 770-788.
doi: 10.1038/s41576-018-0059-1 |
| [27] |
Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature, 2016, 533(7603): 420-424.
doi: 10.1038/nature17946 |
| [28] |
Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, Liu DR. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature, 2017, 551(7681): 464-471.
doi: 10.1038/nature24644 |
| [29] |
Nishida K, Arazoe T, Yachie N, Banno S, Kakimoto M, Tabata M, Mochizuki M, Miyabe A, Araki M, Hara KY, Shimatani Z, Kondo A. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science, 2016, 353(6305): aaf8729.
doi: 10.1126/science.aaf8729 |
| [30] |
Mok BY, de Moraes MH, Zeng J, Bosch DE, Kotrys AV, Raguram A, Hsu F, Radey MC, Peterson SB, Mootha VK, Mougous JD, Liu DR. A bacterial cytidine deaminase toxin enables CRISPR-free mitochondrial base editing. Nature, 2020, 583(7817): 631-637.
doi: 10.1038/s41586-020-2477-4 |
| [31] | Krokan HE, Bjørås M. Base excision repair. Cold Spring Harb Perspect Biol, 2013, 5(4): a012583. |
| [32] |
Aushev M, Herbert M. Mitochondrial Genome Editing Gets Precise. Nature, 2020, 583(7817): 521-522.
doi: 10.1038/d41586-020-01974-6 |
| [33] |
Barrera-Paez JD, Moraes CT. Mitochondrial genome engineering coming-of-age. Trends Genet, 2022, 38(8): 869-880.
doi: 10.1016/j.tig.2022.04.011 pmid: 35599021 |
| [34] | Bi R, Li Y, Xu M, Zheng QZ, Zhang DF, Li X, Ma GL, Xiang BL, Zhu XJ, Zhao H, Huang XX, Zheng P, Yao YG. Direct evidence of CRISPR-Cas9-mediated mitochondrial genome editing. Innovation (Camb), 2022, 3(6): 100329. |
| [35] |
Riepsaame J. Editing the mitochondrial genome: no CRISPR required. Trends Genet, 2020, 36(11): 809-810.
doi: 10.1016/j.tig.2020.08.001 pmid: 32819722 |
| [36] |
Lee H, Lee S, Baek G, Kim A, Kang BC, Seo H, Kim JS. Mitochondrial DNA editing in mice with DddA-TALE fusion deaminases. Nat Commun, 2021, 12(1): 1190.
doi: 10.1038/s41467-021-21464-1 pmid: 33608520 |
| [37] |
Guo JY, Chen XX, Liu ZW, Sun HF, Zhou Y, Dai YC, Ma YE, He L, Qian XZ, Wang JY, Zhang J, Zhu YC, Zhang J, Shen B, Zhou F. DdCBE mediates efficient and inheritable modifications in mouse mitochondrial genome. Mol Ther Nucleic Acids, 2021, 27: 73-80.
doi: 10.1016/j.omtn.2021.11.016 |
| [38] |
Lee S, Lee H, Baek G, Namgung E, Park JM, Kim S, Hong S, Kim JS. Enhanced mitochondrial DNA editing in mice using nuclear-exported TALE-linked deaminases and nucleases. Genome Biol, 2022, 23(1): 211.
doi: 10.1186/s13059-022-02782-z pmid: 36224582 |
| [39] |
Guo JY, Zhang X, Chen XX, Sun HF, Dai YC, Wang JY, Qian XZ, Tan L, Lou X, Shen B. Precision modeling of mitochondrial diseases in zebrafish via DdCBE-mediated mtDNA base editing. Cell Discov, 2021, 7(1): 78.
doi: 10.1038/s41421-021-00307-9 pmid: 34480028 |
| [40] |
Qi XL, Chen XX, Guo JY, Zhang X, Sun HF, Wang JY, Qian XZ, Li B, Tan L, Yu L, Chen W, Zhang LF, Ma YW, Shen B. Precision modeling of mitochondrial disease in rats via DdCBE-mediated mtDNA editing. Cell Discov, 2021, 7(1): 95.
doi: 10.1038/s41421-021-00325-7 pmid: 34663794 |
| [41] |
Chen YP, Lüttmann FF, Schoger E, Schöler HR, Zelarayán LC, Kim KP, Haigh JJ, Kim J, Braun T. Reversible reprogramming of cardiomyocytes to a fetal state drives heart regeneration in mice. Science, 2021, 373(6562): 1537-1540.
doi: 10.1126/science.abg5159 pmid: 34554778 |
| [42] |
Bergmann O, Zdunek S, Felker A, Salehpour M, Alkass K, Bernard S, Sjostrom SL, Szewczykowska M, Jackowska T, Dos Remedios C, Malm T, Andrä M, Jashari R, Nyengaard JR, Possnert G, Jovinge S, Druid H, Frisén J. Dynamics of cell generation and turnover in the human heart. Cell, 2015, 161(7): 1566-1575.
doi: 10.1016/j.cell.2015.05.026 pmid: 26073943 |
| [43] |
Silva-Pinheiro P, Nash PA, Van Haute L, Mutti CD, Turner K, Minczuk M. In vivo mitochondrial base editing via adeno-associated viral delivery to mouse post-mitotic tissue. Nat Commun, 2022, 13(1): 750.
doi: 10.1038/s41467-022-28358-w pmid: 35136065 |
| [44] |
Mok YG, Lee JM, Chung E, Lee J, Lim K, Cho SI, Kim JS. Base editing in human cells with monomeric DddA-TALE fusion deaminases. Nat Commun, 2022, 13(1): 4038.
doi: 10.1038/s41467-022-31745-y pmid: 35821233 |
| [45] |
Chen XX, Liang D, Guo JY, Zhang JQ, Sun HF, Zhang XL, Jin JC, Dai YC, Bao QM, Qian XZ, Tan L, Hu P, Ling XF, Shen B, Xu ZF. DdCBE-Mediated mitochondrial base editing in human 3PN embryos. Cell Discov, 2022, 8(1): 8.
doi: 10.1038/s41421-021-00358-y pmid: 35102135 |
| [46] |
St John JC, Facucho-Oliveira J, Jiang Y, Kelly R, Salah R. Mitochondrial DNA transmission, replication and inheritance: a journey from the gamete through the embryo and into offspring and embryonic stem cells. Hum Reprod Update, 2010, 16(5): 488-509.
doi: 10.1093/humupd/dmq002 pmid: 20231166 |
| [47] |
Wei YH, Xu CL, Feng H, Xu K, Li ZF, Hu J, Zhou L, Wei Y, Zuo ZR, Zuo EW, Li W, Yang H, Zhang ML. Human cleaving embryos enable efficient mitochondrial base- editing with DdCBE. Cell Discov, 2022, 8(1): 7.
doi: 10.1038/s41421-021-00372-0 |
| [48] |
Wrighton KH. Cytosine base editors go off-target. Nat Rev Genet, 2019, 20(5): 254-255.
doi: 10.1038/s41576-019-0110-x pmid: 30872767 |
| [49] |
Zuo EW, Sun YD, Wei W, Yuan TL, Ying WQ, Sun H, Yuan LY, Steinmetz LM, Li YX, Yang H. GOTI, a method to identify genome-wide off-target effects of genome editing in mouse embryos. Nat Protoc, 2020, 15(9): 3009-3029.
doi: 10.1038/s41596-020-0361-1 pmid: 32796939 |
| [50] |
Zuo EW, Sun YD, Wei W, Yuan TL, Ying WQ, Sun H, Yuan LY, Steinmetz LM, Li YX, Yang H. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science, 2019, 364(6437): 289-292.
doi: 10.1126/science.aav9973 pmid: 30819928 |
| [51] |
Wei YH, Li ZF, Xu K, Feng H, Xie L, Li D, Zuo ZR, Zhang ML, Xu CL, Yang H, Zuo EW. Mitochondrial base editor DdCBE causes substantial DNA off-target editing in nuclear genome of embryos. Cell Discov, 2022, 8(1): 27.
doi: 10.1038/s41421-022-00391-5 pmid: 35304438 |
| [52] | Lei ZX, Meng HW, Lv ZC, Liu MH, Zhao HN, Wu H, Zhang XX, Liu LL, Zhuang Y, Yin KL, Yan YC, Yi CQ. Detect-seq reveals out-of-protospacer editing and target- strand editing by cytosine base editors. Nat Methods, 2021, 18(6): 643-651. |
| [53] |
Lei ZX, Meng HW, Liu LL, Zhao HN, Rao XC, Yan YC, Wu H, Liu M, He AB, Yi CQ. Mitochondrial base editor induces substantial nuclear off-target mutations. Nature, 2022, 606(7915): 804-811.
doi: 10.1038/s41586-022-04836-5 |
| [54] |
Rowley MJ, Corces VG. Organizational principles of 3D genome architecture. Nat Rev Genet, 2018, 19(12): 789-800.
doi: 10.1038/s41576-018-0060-8 pmid: 30367165 |
| [55] |
Hansen AS, Pustova I, Cattoglio C, Tjian R, Darzacq X. CTCF and cohesin regulate chromatin loop stability with distinct dynamics. eLife, 2017, 6: e25776.
doi: 10.7554/eLife.25776 |
| [56] |
Lee S, Lee H, Baek G, Kim JS. Precision mitochondrial DNA editing with high-fidelity DddA-derived base editors. Nat Biotechnol, 2023, 41(3): 378-386.
doi: 10.1038/s41587-022-01486-w |
| [57] |
Miller JC, Holmes MC, Wang JB, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, Gregory PD, Pabo CO, Rebar EJ. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol, 2007, 25(7): 778-785.
doi: 10.1038/nbt1319 pmid: 17603475 |
| [58] |
Szczepek M, Brondani V, Büchel J, Serrano L, Segal DJ, Cathomen T. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol, 2007, 25(7): 786-793.
doi: 10.1038/nbt1317 pmid: 17603476 |
| [59] |
Mok BY, Kotrys AV, Raguram A, Huang TP, Mootha VK, Liu DR. CRISPR-free base editors with enhanced activity and expanded targeting scope in mitochondrial and nuclear DNA. Nat Biotechnol, 2022, 40(9): 1378-1387.
doi: 10.1038/s41587-022-01256-8 pmid: 35379961 |
| [60] |
Thuronyi BW, Koblan LW, Levy JM, Yeh WH, Zheng C, Newby GA, Wilson C, Bhaumik M, Shubina-Oleinik O, Holt JR, Liu DR. Continuous evolution of base editors with expanded target compatibility and improved activity. Nat Biotechnol, 2019, 37(9): 1070-1079.
doi: 10.1038/s41587-019-0193-0 pmid: 31332326 |
| [61] |
Roth TB, Woolston BM, Stephanopoulos G, Liu DR.Phage-assisted evolution of bacillus methanolicus methanol dehydrogenase 2. Acs Synth Biol, 2019, 8(4): 796-806.
doi: 10.1021/acssynbio.8b00481 pmid: 30856338 |
| [62] |
Dickinson BC, Packer MS, Badran AH, Liu DR. A system for the continuous directed evolution of proteases rapidly reveals drug-resistance mutations. Nat Commun, 2014, 5(1): 5352.
doi: 10.1038/ncomms6352 |
| [63] |
Cho SI, Lee S, Mok YG, Lim K, Lee J, Lee JM, Chung E, Kim JS. Targeted A-to-G base editing in human mitochondrial DNA with programmable deaminases. Cell, 2022, 185(10): 1764-1776.e12.
doi: 10.1016/j.cell.2022.03.039 |
| [64] |
Richter MF, Zhao KT, Eton E, Lapinaite A, Newby GA, Thuronyi BW, Wilson C, Koblan LW, Zeng J, Bauer DE, Doudna JA, Liu DR. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat Biotechnol, 2020, 38(7): 883-891.
doi: 10.1038/s41587-020-0453-z pmid: 32433547 |
| [65] | Morgan MA, Lange L, Schambach A. Prime time for base editing in the mitochondria. Signal Transduct Target Ther, 2022, 7(1): 213. |
| [66] |
Willis JCW, Silva-Pinheiro P, Widdup L, Minczuk M, Liu DR. Compact zinc finger base editors that edit mitochondrial or nuclear DNA in vitro and in vivo. Nat Commun, 2022, 13(1): 7204.
doi: 10.1038/s41467-022-34784-7 pmid: 36418298 |
| [67] |
Lim K, Cho SI, Kim JS. Nuclear and mitochondrial DNA editing in human cells with zinc finger deaminases. Nat Commun, 2022, 13(1): 366.
doi: 10.1038/s41467-022-27962-0 pmid: 35042880 |
| [68] |
Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, Chen PJ, Wilson C, Newby GA, Raguram A, Liu DR. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature, 2019, 576(7785): 149-157.
doi: 10.1038/s41586-019-1711-4 |
| [69] |
Chen PJ, Liu DR. Prime editing for precise and highly versatile genome manipulation. Nat Rev Genet, 2023, 24(3): 161-177.
doi: 10.1038/s41576-022-00541-1 |
| [70] | Zong Y, Gao CX. Progress on base editing systems. Hereditas(Beijing), 2019, 41(9): 777-800. |
| 宗媛, 高彩霞. 碱基编辑系统研究进展. 遗传, 2019, 41(9): 777-800. |
| [1] | 胡风越, 王克剑. STEME系统:一种助力体内定向进化的新工具[J]. 遗传, 2020, 42(3): 231-235. |
| [2] | 宗媛, 高彩霞. 碱基编辑系统研究进展[J]. 遗传, 2019, 41(9): 777-800. |
| [3] | 任云晓, 肖茹丹, 娄晓敏, 方向东. 基因编辑技术及其在基因治疗中的应用[J]. 遗传, 2019, 41(1): 18-27. |
| [4] | 魏瑜,张晓辉,李大力. 基因编辑之“新宠”—单碱基基因组编辑系统[J]. 遗传, 2017, 39(12): 1115-1121. |
| [5] | 许美芬, 何轶群, 耿军伟, 孟燕子, 于涵, 林枝, 施苏雪, 薛凌, 卢中秋, 管敏鑫. 两个携带线粒体tRNAMet/tRNAGlnA4401G和tRNACysG5821A突变的中国汉族原发性高血压家系的临床及分子遗传学特征[J]. 遗传, 2014, 36(2): 127-134. |
| [6] | 海萨·艾也力汗,郭焱,孟玮,杨天燕,马燕武. 新疆裂腹鱼类的系统发生关系及物种分化时间[J]. 遗传, 2014, 36(10): 1013-1020. |
| [7] | 张初琴,陈波蓓,陈迎迎,刘学军,郑静,高金建,黄赛瑜,南奔宇,章誉耀,余啸,管敏鑫. 不同年龄段非综合征性耳聋常见基因检测及临床表型分析[J]. 遗传, 2013, 35(3): 352-358. |
| [8] | 马志杰,钟金城,韩建林,徐惊涛,刘仲娜,白文林. 牦牛分子遗传多样性研究进展[J]. 遗传, 2013, 35(2): 151-160. |
| [9] | 张阿梅 姚永刚. Leber遗传性视神经病变研究进展和挑战[J]. 遗传, 2013, 35(2): 123-135. |
| [10] | 王金凤,张亚平,于黎. 食肉目猫科物种的系统发育学研究概述[J]. 遗传, 2012, 34(11): 1365-1378. |
| [11] | 冀延春,刘晓玲,赵福新,张娟娟,章豫,周翔天,瞿佳,管敏鑫. 线粒体T12338C突变可能是与Leber遗传性视神经病变相关的突变位点[J]. 遗传, 2011, 33(4): 322-328. |
| [12] | 危金普,潘学峰,李红权,段斐. 简单重复DNA序列在哺乳动物mtDNA D-loop区的分布及进化特征[J]. 遗传, 2011, 33(1): 67-74. |
| [13] | 张永梅,冀延春,刘晓玲,周翔天,赵福新,孙艳红,韦企平,张娟娟,刘燕,瞿佳,管敏鑫. 线粒体tRNA A14693G可能是与Leber遗传性视神经病变相关的基因突变[J]. 遗传, 2010, 32(4): 353-359. |
| [14] | 刘燕,庄淑流,童绎,瞿佳,周翔天,赵福新,张娟娟,张永梅,章豫. 线粒体ND1基因T3866C突变可能是Leber's遗传性 视神经病和四肢畸形跛行相关的突变[J]. 遗传, 2010, 32(2): 141-147. |
| [15] | 杨爱芬,朱翌,吕建新,杨丽,赵建越,孙冬梅,管敏鑫. 线粒体DNA G7444A突变可能影响A1555G突变的表型表达[J]. 遗传, 2008, 30(6): 728-734. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||

www.chinagene.cn
备案号:京ICP备09063187号-4
总访问:,今日访问:,当前在线:
