遗传 ›› 2025, Vol. 47 ›› Issue (8): 842-860.doi: 10.16288/j.yczz.24-311
周迅1,2,3( ), 周世杰1,2,3, 刘捷1,2,3, 王宇祥1,2,3(
), 周世杰1,2,3, 刘捷1,2,3, 王宇祥1,2,3( )
)
收稿日期:2024-11-01
修回日期:2025-02-23
出版日期:2025-08-20
发布日期:2025-04-27
通讯作者:
王宇祥,教授,研究方向:动物分子遗传学。E-mail: wyx2000@neau.edu.cn作者简介:周迅,硕士研究生,专业方向:动物分子遗传学。E-mail: s230502025@neau.edn.cn
基金资助:
Xun Zhou1,2,3( ), Shijie Zhou1,2,3, Jie Liu1,2,3, Yuxiang Wang1,2,3(
), Shijie Zhou1,2,3, Jie Liu1,2,3, Yuxiang Wang1,2,3( )
)
Received:2024-11-01
Revised:2025-02-23
Published:2025-08-20
Online:2025-04-27
Supported by:摘要:
RNA编辑是表观遗传学领域重要的研究方向之一。随着研究的深入,科学家们发现CRISPR/Cas系统不仅可以靶向DNA,也可以靶向RNA,从而实现转录水平的基因精准编辑;同时,使用CRISPR/Cas系统进行RNA编辑也可以避免对基因组的破坏。目前,基于靶向RNA的CRISPR系统已开发出多种衍生技术,如RNA敲低和编辑、核酸检测和成像、RNA示踪等。这些衍生技术的出现,为生物遗传机制解析和疾病治疗提供了有利工具。本文归纳总结了靶向RNA的CRISPR/Cas系统的结构、功能、机制以及开发的衍生技术,以期丰富人们对CRISPR/Cas系统编辑RNA的认知。
周迅, 周世杰, 刘捷, 王宇祥. 靶向RNA的CRISPR/Cas系统及衍生技术[J]. 遗传, 2025, 47(8): 842-860.
Xun Zhou, Shijie Zhou, Jie Liu, Yuxiang Wang. CRISPR/Cas system targeting RNA and its derivative technology[J]. Hereditas(Beijing), 2025, 47(8): 842-860.
表1
CRISPR/Cas系统分类"
| 大类 | 型 | 亚型 | 包含的Cas蛋白 | 主要靶向的目标 |
|---|---|---|---|---|
| 第一类 | I型 | I-A、I-B、I-C、I-D、I-E、I-F1、I-F2、I-F3 | Cas1、Cas2、Cas3、Cas4、Cas5、Cas6、Cas7、Cas8 | ssDNA |
| III型 | III-A、III-B、III-C、III-D、III-E、III-F | Cas1、Cas2、Cas5、Cas6、Cas7、Cas10、Cas11 | ssDNA和ssRNA | |
| IV型 | IV-A、IV-B、IV-C | Cas1、Cas2、Cas5、Cas6、Cas7 | — | |
| VII型(候选成员) | — | Cas5、Cas7、Cas6、Cas14 | ssRNA | |
| 第二类 | II型 | II-A、II-B、II-C1、II-C2(效应蛋白都称为Cas9) | Cas1、Cas2、Cas4、Cas9 | dsDNA |
| V型 | V-A(Cas12a)、V-B(Cas12b)、V-C(Cas12c)、V-D(Cas12d)、V-E(Cas13e)、V-F1(Cas14a)、V-U3(C2c10)、V-F2(Cas14b)、V-F3(Cas14c)、V-G(Cas12g)、V-U1(C2c4)、V-U2(C2c8)、V-U4(C2c9)、V-K/V-U5(C2c5) | Cas1、Cas2、Cas4、Cas12 | ssDNA和dsDNA | |
| VI型 | VI-A(Cas13a)、VI-B1(Cas1b1)、VI-B2(Cas13b2)、VI-C(Cas13c)、VI-D(Cas13d) | Cas1、Cas2、Cas13 | ssRNA |
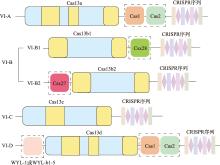
图2
VI型CRISPR/Cas系统结构示意图 VI-A和VI-D系统包含编码Cas1和Cas2蛋白的基因,VI-B系统包含翻译Csx28和Csx27辅助蛋白的基因,根据辅助蛋白的不同又可细分为VI-B1和VI-B2两类,其中VI-B1中的辅助蛋白Csx28起促进Cas13b的作用,VI-B2的辅助蛋白Csx27起抑制Cas13b的作用;VI-D系统中包含具有WYL结构域(以保守的Trp-Tyr-Leu基序命名)的辅助蛋白WYL-1或WYL-b1-5的基因。Cas1、Cas2、Csx27、Csx28、WYL-1和WYL-b1-5结构(图中虚线框中的蛋白)只存在于拥有特定亚型的某些物种中。"
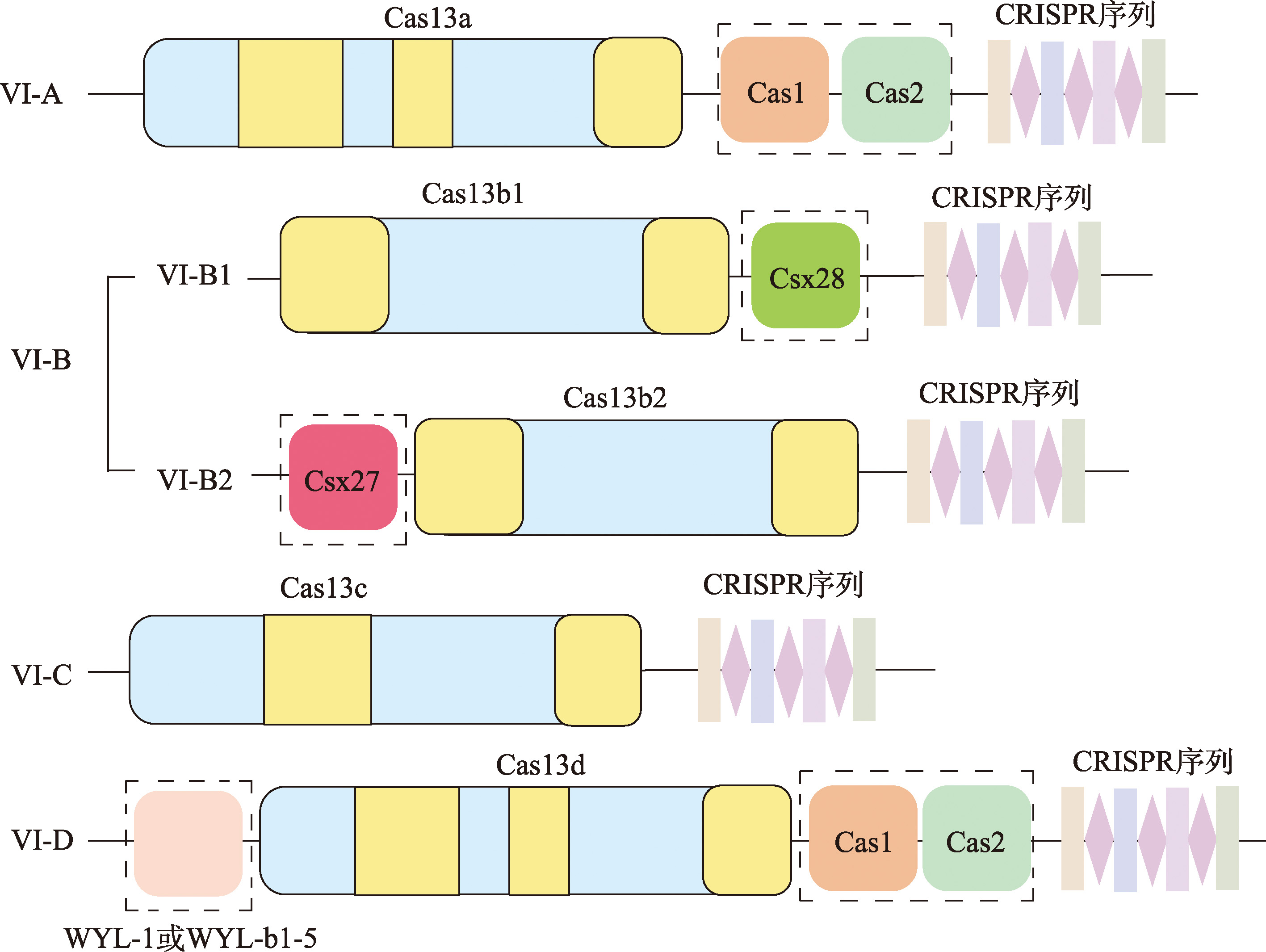
表2
不同III型系统的复合物类型及功能"
| 分类 | 复合物 类型 | 特异性 ssRNase 活性 | ssDNase 活性 | 合成cOAs (造成非特 异性RNA 切割) | 核心 Cas蛋白 | Csm/Cmr复合物的组成 | 共同的 Cas蛋白 | 不同的 Cas蛋白 |
|---|---|---|---|---|---|---|---|---|
| III-A | Csm | 是 | 是 | 是 | Cas10 | Cas5(Csm4)、Cas7(Csm3/Csm5)、Cas10(Csm1)、Cas11(Csm2) | 除III-E系统只有Cas7和Cas11外,其他5个III型系统都包含Cas5、Cas7、Cas10和Cas11这4个Cas蛋白 | Cas1、Cas2 每个亚型携带的辅助蛋白 |
| III-B | Cmr | 是 | 是 | 是 | Cas10 | Cas5(Cmr3)、Cas7(Cmr4/Cmr1/Cmr6)、Cas10(Cmr2)、Cas11(Cmr5) | ||
| III-C | Cmr | 是 | 是 | 否 | Cas10 | Cas5(Cmr3)、Cas7(Cmr4/Cmr1/Cmr6)、Cas10(Cmr2)、Cas11(Cmr5) | ||
| III-D | Csm | 是 | 否 | 是 | Cas10 | Cas5(Csm4)、Cas7(Csm3/Csm5)、Cas10(Csm1)、Cas11(Csm2) | ||
| III-E | Csm | 是 | 否 | 否 | Cas7和 Cas11 | Cas7和Cas11 | ||
| III-F | Csm | 否 | 是 | 是 | Cas10 | Cas5(Csm4)、Cas7(Csm3)、 Cas10(Csm1)、Cas11(Csm2) |
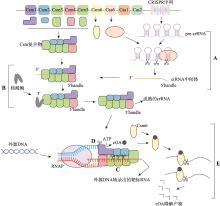
图6
III型CRISPR/Cas系统机制图 III型系统行使功能具体机制:首先Cas6加工pre-crRNA,形成crRNA中间体(A);之后,在核酸酶的帮助下,对crRNA中间体3′末端进行剪切处理,形成成熟的crRNA(B);当靶标RNA被转录出来后,crRNA识别并结合靶标RNA,由Csm复合物中的Cas7剪切靶标RNA(C);如果转录出的靶标RNA不含有重复序列,则crRNA的5′手柄不与靶标RNA互补结合,Cas10蛋白HD结构域的单链DNase活性被激活,剪切非模板ssDNA(D);同时,crRNA 5′手柄未与靶标RNA互补结合,还会激活Cas10蛋白Plam结构域的环化酶活性,使ATP转化为cOA;cOA再与Csm6蛋白的CARF结构域结合,激活Csm6蛋白的RNase活性,剪切周围的非特异性RNA(E);之后,Csm6还可行使环状核酸酶的功能,降解cOA抑制或关闭该过程。CRISPR序列中长方形代表间隔序列,菱形代表重复序列;图中Csm复合物中方块颜色与Cas基因簇中图形颜色相同的表示为同一蛋白;图中Csm复合物为简图,具体结构见图5。RNAP:RNA聚合酶(RNA polymerase)。"
| [1] |
Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science, 2007, 315(5819): 1709-1712.
pmid: 17379808 |
| [2] |
Shabalina SA, Koonin EV. Origins and evolution of eukaryotic RNA interference. Trends Ecol Evol, 2008, 23(10): 578-587.
pmid: 18715673 |
| [3] |
Hur JK, Olovnikov I, Aravin AA. Prokaryotic argonautes defend genomes against invasive DNA. Trends Biochem Sci, 2014, 39(6): 257-259.
pmid: 24836995 |
| [4] |
Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol, 2008, 9(1): 22-32.
pmid: 18073770 |
| [5] |
Koonin EV, Makarova KS. Origins and evolution of CRISPR-cas systems. Philos Trans R Soc Lond B Biol Sci, 2019, 374(1772): 20180087.
pmid: 30905284 |
| [6] |
Koonin EV, Makarova KS. CRISPR-Cas: an adaptive immunity system in prokaryotes. F1000 Biol Rep, 2009, 1: 95.
pmid: 24766887 |
| [7] |
Nidhi S, Anand U, Oleksak P, Tripathi P, Lal JA, Thomas G, Kuca K, Tripathi V. Novel CRISPR-cas systems: an updated review of the current achievements, applications, and future research perspectives. Int J Mol Sci, 2021, 22(7): 3327.
pmid: 33805113 |
| [8] |
Alkhnbashi OS, Shah SA, Garrett RA, Saunders SJ, Costa F, Backofen R. Characterizing leader sequences of CRISPR loci. Bioinformatics, 32(17): i576-i585.
pmid: 27587677 |
| [9] |
Makarova KS, Wolf YI, Iranzo J, Shmakov SA, Alkhnbashi OS, Brouns SJJ, Charpentier E, Cheng D, Haft DH, Horvath P, Moineau S, Mojica FJM, Scott D, Shah SA, Siksnys V, Terns MP, Venclovas Č, White MF, Yakunin AF, Yan W, Zhang F, Garrett RA, Backofen R, Van Der Oost J, Barrangou R, Koonin EV. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol, 2020, 18(2): 67-83.
pmid: 31857715 |
| [10] | Liu Y, Zhang SF, Hu CY. Cas7 meets Cas14: a strategic partnership in the type VII CRISPR-Cas. Protein Cell, 2024, 16(2): 79-82. |
| [11] | Gao WS, Dou JP, Wei S, Liu XJ, Zhang ZF, Li YN. Classification and research status of CRISPR/Cas systems. Biotechnol Progr, 2022, 12(4): 532-538. |
| 高维崧, 窦金萍, 韦双, 刘兴健, 张志芳, 李轶女. CRISPR/Cas系统的分类及研究现状. 生物技术进展, 2022, 12(4): 532-538. | |
| [12] |
Amitai G, Sorek R. CRISPR-Cas adaptation: insights into the mechanism of action. Nat Rev Microbiol, 2016, 14(2): 67-76
pmid: 26751509 |
| [13] |
Jackson SA, McKenzie RE, Fagerlund RD, Kieper SN, Fineran PC, Brouns SJJ. CRISPR-Cas: adapting to change. Science, 2017, 356(6333): eaal5056
pmid: 28385959 |
| [14] |
Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJM, Terns RM, Terns MP, White MF, Yakunin AF, Garrett RA, Van Der Oost J, Backofen R, Koonin EV. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol, 2015, 13(11): 722-736.
pmid: 26411297 |
| [15] |
Mohanraju P, Makarova KS, Zetsche B, Zhang F, Koonin EV. Van Der Oost J. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science, 2016, 353(6299): aad5147.
pmid: 27493190 |
| [16] |
Makarova KS H, Haft DH, Barrangou R, Brouns SJJ, Charpentier E, Horvath P, Moineau S, Mojica FJM, Wolf YI, Yakunin AF, Oost JVD, Koonin EV. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol, 2011, 9(6): 467-477.
pmid: 21552286 |
| [17] |
Hillary VE, Ceasar SA. A review on the mechanism and applications of CRISPR/Cas9/Cas12/Cas13/Cas14 proteins utilized for genome engineering. Mol Biotechnol, 2023, 65(3): 311-325.
pmid: 36163606 |
| [18] |
Koonin EV, Makarova KS, Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol, 2017, 37: 67-78.
pmid: 28605718 |
| [19] |
Burmistrz M, Krakowski K, Krawczyk-Balska A. RNA-targeting CRISPR-Cas systems and their applications. Int J Mol Sci, 2020, 21(3): 1122.
pmid: 32046217 |
| [20] |
Shmakov S, Smargon A, Scott D, Cox D, Pyzocha N, Yan W, Abudayyeh OO, Gootenberg JS, Makarova KS, Wolf YI, Severinov K, Zhang F, Koonin EV. Diversity and evolution of class 2 CRISPR-Cas systems. Nat Rev Microbiol, 2017, 15(3): 169-182.
pmid: 28111461 |
| [21] |
Kavuri NR, Ramasamy M, Qi YP, Mandadi K. Applications of CRISPR/Cas13-based RNA editing in plants. Cells, 2022, 11(17): 2665.
pmid: 36078073 |
| [22] |
Yang H, Patel DJ. Structures, mechanisms and applications of RNA-centric CRISPR-Cas13. Nat Chem Biol, 2024, 20(6): 673-688.
pmid: 38702571 |
| [23] |
Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DBT, Shmakov S, Makarova KS, Semenova E, Minakhin L, Severinov K, Regev A, Lander ES, Koonin EV, Zhang F. C2c2 is a single- component programmable RNA-guided RNA-targeting CRISPR effector. Science, 2016, 353(6299): aaf5573.
pmid: 27256883 |
| [24] |
Smargon AA, Cox DBT, Pyzocha NK, Zheng KJ, Slaymaker IM, Gootenberg JS, Abudayyeh OA, Essletzbichler P, Shmakov S, Makarova KS, Koonin EV, Zhang F. Cas13b is a type VI-B CRISPR-associated RNA-guided RNase differentially regulated by accessory proteins Csx27 and Csx28. Mol Cell, 2017, 65(4): 618-630.e7.
pmid: 28065598 |
| [25] |
Yan WX, Chong SR, Zhang HB, Makarova KS, Koonin EV, Cheng DR, Scott DA. Cas13d is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-Domain-Containing accessory protein. Mol Cell, 2018, 70(2): 327-339.e5.
pmid: 29551514 |
| [26] |
Hu YP, Chen YC, Xu J, Wang XG, Luo SQ, Mao BW, Zhou Q, Li W. Metagenomic discovery of novel CRISPR- Cas13 systems. Cell Discov, 2022, 8(1): 107.
pmid: 36220832 |
| [27] |
Xu CL, Zhou YS, Xiao QQ, He BB, Geng GN, Wang ZK, Cao BR, Dong X, Bai WY, Wang YF, Wang X, Zhou DM, Yuan TL, Huo XN, Lai JS, Yang H. Programmable RNA editing with compact CRISPR-Cas13 systems from uncultivated microbes. Nat Methods, 2021, 18(5): 499-506.
pmid: 33941935 |
| [28] |
Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, Minakhin L, Joung J, Konermann S, Severinov K, Zhang F, Koonin EV. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol Cell, 2015, 60(3): 385-397.
pmid: 26593719 |
| [29] |
Konermann S, Lotfy P, Brideau NJ, Oki J, Shokhirev MN, Hsu PD. Transcriptome engineering with RNA- targeting type VI-D CRISPR effectors. Cell, 2018, 173(3): 665-676.e14.
pmid: 29551272 |
| [30] |
Perčulija V, Lin JY, Zhang B, Ouyang SY. Functional features and current applications of the RNA-targeting type VI CRISPR-Cas systems. Adv Sci (Weinh), 2021, 8(13): 2004685.
pmid: 34254038 |
| [31] |
Liu L, Li XY, Wang JY, Wang M, Chen P, Yin ML, Li JZ, Sheng G, Wang YL. Two distant catalytic sites are responsible for C2c2 RNase activities. Cell, 2017, 168(1-2): 121-134.e12.
pmid: 28086085 |
| [32] |
Liu L, Li XY, Ma J, Li ZQ, You LL, Wang JY, Wang M, Zhang XZ, Wang YL. The molecular architecture for RNA-guided RNA cleavage by Cas13a. Cell, 2017, 170(4): 714-726.e10.
pmid: 28757251 |
| [33] |
Knott GJ, East-Seletsky A, Cofsky JC, Holton JM, Charles E, O’Connell MR, Doudna JA. Guide-bound structures of an RNA-targeting A-cleaving CRISPR- Cas13a enzyme. Nat Struct Mol Biol, 2017, 24(10): 825-833.
pmid: 28892041 |
| [34] |
Zhang B, Ye WW, Ye YM, Zhou H, Saeed AFUH, Chen J, Lin JY, Perčulija V, Chen Q, Chen CJ, Chang MX, Choudhary MI, Ouyang SY. Structural insights into Cas13b-guided CRISPR RNA maturation and recognition. Cell Res, 2018, 28(12): 1198-1201.
pmid: 30425321 |
| [35] |
Slaymaker IM, Mesa P, Kellner MJ, Kannan S, Brignole E, Koob J, Feliciano PR, Stella S, Abudayyeh OO, Gootenberg JS, Strecker J, Montoya G, Zhang F. High- resolution structure of Cas13b and biochemical characterization of RNA targeting and cleavage. Cell Rep, 2019, 26(13): 3741-3751.e5.
pmid: 30917325 |
| [36] |
Zhang C, Konermann S, Brideau NJ, Lotfy P, Wu XB, Novick SJ, Strutzenberg T, Griffin PR, Hsu PD, Lyumkis D. Structural basis for the RNA-guided ribonuclease activity of CRISPR-Cas13d. Cell, 2018, 175(1): 212-223.e17.
pmid: 30241607 |
| [37] |
O’Connell MR. Molecular mechanisms of RNA targeting by Cas13-containing type VI CRISPR-Cas systems. J Mol Biol, 2019, 431(1): 66-87.
pmid: 29940185 |
| [38] |
Zhang B, Ye YM, Ye WW, Perčulija V, Jiang H, Chen YY, Li Y, Chen J, Lin JY, Wang SQ, Chen Q, Han YS, Ouyang SY. Two HEPN domains dictate CRISPR RNA maturation and target cleavage in Cas13d. Nat Commun, 2019, 10(1): 2544.
pmid: 31186424 |
| [39] |
East-Seletsky A, O’Connell MR, Burstein D, Knott GJ, Doudna JA. RNA targeting by functionally orthogonal type VI-A CRISPR-Cas enzymes. Mol Cell, 2017, 66(3): 373-383.e3.
pmid: 28475872 |
| [40] |
Li XZC, Han J, Yang J, Zhang H. The structural biology of type III CRISPR-Cas systems. J Struct Biol, 2024, 216(1): 108070.
pmid: 38395113 |
| [41] |
Kolesnik MV, Fedorova I, Karneyeva KA, Artamonova DN, Severinov KV. Type III CRISPR-Cas systems: deciphering the most complex prokaryotic immune system. Biochemistry (Mosc), 2021, 86(10): 1301-1314.
pmid: 34903162 |
| [42] |
Hamdi I, Boni F, Shen QL, Moukendza L, Li PB, Xie JP. Characteristics of subtype III-a CRISPR-cas system in mycobacterium tuberculosis: an overview. Infect Genet Evol, 2023, 112: 105445.
pmid: 37217031 |
| [43] |
Molina R, Sofos N, Montoya G. Structural basis of CRISPR-Cas type III prokaryotic defence systems. Curr Opin Struct Biol, 2020, 65: 119-129.
pmid: 32712502 |
| [44] |
Kazlauskiene M, Tamulaitis G, Kostiuk G, Venclovas Č, Siksnys V. Spatiotemporal control of type III-A CRISPR- Cas immunity: coupling DNA degradation with the target RNA recognition. Mol Cell, 2016, 62(2): 295-306.
pmid: 27105119 |
| [45] |
Özcan A, Krajeski R, Ioannidi E, Lee B, Gardner A, Makarova KS, Koonin EV, Abudayyeh OO, Gootenberg JS. Programmable RNA targeting with the single-protein CRISPR effector Cas7-11. Nature, 2021, 597(7878): 720-725.
pmid: 34489594 |
| [46] |
Steens JA, van der Oost J, Staals RHJ. Compact but mighty: biology and applications of type III-E CRISPR- Cas systems. Mol Cell, 2022, 82(23): 4405-4406.
pmid: 36459983 |
| [47] |
Bhatia S, Pooja, Yadav SK. CRISPR-Cas for genome editing: classification, mechanism, designing and applications. Int J Biol Macromol, 2023, 238: 124054.
pmid: 36933595 |
| [48] |
Samai P, Pyenson N, Jiang WY, Goldberg GW, Hatoum- Aslan A, Marraffini LA. Co-transcriptional DNA and RNA cleavage during type III CRISPR-Cas immunity. Cell, 2015, 161(5): 1164-1174.
pmid: 25959775 |
| [49] |
Staals RHJ, Zhu YF, Taylor DW, Kornfeld JE, Sharma K, Barendregt A, Koehorst JJ, Vlot M, Neupane N, Varossieau K, Sakamoto K, Suzuki T, Dohmae N, Yokoyama S, Schaap PJ, Urlaub H, Heck AJR, Nogales E, Doudna JA, Shinkai A, van der Oost J. RNA targeting by the type III-A CRISPR-Cas Csm complex of Thermus thermophilus. Mol Cell, 2014, 56(4): 518-530.
pmid: 25457165 |
| [50] |
Ramia NF, Spilman M, Tang L, Shao YM, Elmore J, Hale C, Cocozaki A, Bhattacharya N, Terns RM, Terns MP, Li H, Stagg SM. Essential structural and functional roles of the Cmr4 subunit in RNA cleavage by the Cmr CRISPR-Cas complex. Cell Rep, 2014, 9(5): 1610-1617.
pmid: 25482566 |
| [51] |
Peng WF, Feng MX, Feng X, Liang YX, She QX. An archaeal CRISPR type III-B system exhibiting distinctive RNA targeting features and mediating dual RNA and DNA interference. Nucleic Acids Res, 2015, 43(1): 406-417.
pmid: 25505143 |
| [52] |
Tamulaitis G, Kazlauskiene M, Manakova E, Venclovas Č, Nwokeoji AO, Dickman MJ, Horvath P, Siksnys V. Programmable RNA shredding by the type III-A CRISPR-Cas system of Streptococcus thermophilus. Mol Cell, 2014, 56(4): 506-517.
pmid: 25458845 |
| [53] |
Estrella MA, Kuo FT, Bailey S. RNA-activated DNA cleavage by the type III-B CRISPR-Cas effector complex. Genes Dev, 2016, 30(4): 460-470.
pmid: 26848046 |
| [54] |
Elmore JR, Sheppard NF, Ramia N, Deighan T, Li H, Terns RM, Terns MP. Bipartite recognition of target RNAs activates DNA cleavage by the type III-B CRISPR-Cas system. Genes Dev, 2016, 30(4): 447-459.
pmid: 26848045 |
| [55] |
Tamulaitis G, Venclovas Č, Siksnys V. Type III CRISPR- Cas immunity: major differences brushed aside. Trends Microbiol, 2017, 25(1): 49-61.
pmid: 27773522 |
| [56] |
Niewoehner O, Garcia-Doval C, Rostøl JT, Berk C, Schwede F, Bigler L, Hall J, Marraffini LA, Jinek M. Type III CRISPR-Cas systems produce cyclic oligoadenylate second messengers. Nature, 2017, 548(7669): 543-548.
pmid: 28722012 |
| [57] |
Kazlauskiene M, Kostiuk G, Venclovas Č, Tamulaitis G, Siksnys V. A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science, 2017, 357(6351): 605-609.9.
pmid: 28663439 |
| [58] |
Niewoehner O, Jinek M. Structural basis for the endoribonuclease activity of the type III-A CRISPR- associated protein Csm6. RNA, 2016, 22(3): 318-329.
pmid: 26763118 |
| [59] |
Shah SZ, Rehman A, Nasir H, Asif A, Tufail B, Usama M, Jabbar B. Advances in research on genome editing CRISPR-Cas9 technology. J Ayub Med Coll Abbottabad, 2019, 31(1): 108-122.
pmid: 30868795 |
| [60] |
Zhu YM. Advances in CRISPR/Cas9. Biomed Res Int, 2022, 2022: 9978571.
pmid: 36193328 |
| [61] |
Strutt SC, Torrez RM, Kaya E, Negrete OA, Doudna JA. RNA-dependent RNA targeting by CRISPR-Cas9. eLife, 2018, 7: e32724.
pmid: 29303478 |
| [62] |
Dugar G, Leenay RT, Eisenbart SK, Bischler T, Aul BU, Beisel CL, Sharma CM. CRISPR RNA-dependent binding and cleavage of endogenous RNAs by the Campylobacter jejuni Cas9. Mol Cell, 2018, 69(5): 893-905.e7.
pmid: 29499139 |
| [63] |
Rousseau BA, Hou ZG, Gramelspacher MJ, Zhang Y. Programmable RNA cleavage and recognition by a natural CRISPR-Cas9 system from Neisseria meningitidis. Mol Cell, 2018, 69(5): 906-914.e4.
pmid: 29456189 |
| [64] |
Gunitseva N, Evteeva M, Borisova A, Patrushev M, Subach F. RNA-dependent RNA targeting by CRISPR- Cas systems: characterizations and applications. Int J Mol Sci, 2023, 24(8): 6894.
pmid: 37108063 |
| [65] |
O’Connell MR, Oakes BL, Sternberg SH, East-Seletsky A, Kaplan M, Doudna JA. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature, 2014, 516(7530): 263-266.
pmid: 25274302 |
| [66] |
Nelles DA, Fang MY, O’Connell MR, Xu JL, Markmiller SJ, Doudna JA, Yeo GW. Programmable RNA tracking in live cells with CRISPR/Cas9. Cell, 2016, 165(2): 488-496.
pmid: 25274302 |
| [67] |
Liu YC, Chen ZC, He AB, Zhan YH, Li JF, Liu L, Wu HW, Zhuang CL, Lin JH, Zhang QX, Huang WR. Targeting cellular mRNAs translation by CRISPR-Cas9. Sci Rep, 2016, 6: 29652.
pmid: 27405721 |
| [68] |
Batra R, Nelles DA, Pirie E, Blue SM, Marina RJ, Wang H, Chaim IA, Thomas JD, Zhang N, Nguyen V, Aigner S, Markmiller S, Xia GB, Corbett KD, Swanson MS, Yeo GW. Elimination of toxic microsatellite repeat expansion RNA by RNA-targeting Cas9. Cell, 2017, 170(5): 899-912.e10.
pmid: 28803727 |
| [69] |
Yan WX, Hunnewell P, Alfonse LE, Carte JM, Keston-Smith E, Sothiselvam S, Garrity AJ, Chong SR, Makarova KS, Koonin EV, Cheng DR, Scott DA. Functionally diverse type V CRISPR-Cas systems. Science, 2019, 363(6422): 88-91.
pmid: 30523077 |
| [70] |
Liu MX, Li ZK, Chen J, Lin JY, Lu QH, Ye YM, Zhang HM, Zhang B, Ouyang SY. Structural transitions upon guide RNA binding and their importance in Cas12g- mediated RNA cleavage. PLoS Genet, 2023, 19(9): e1010930.
pmid: 37729124 |
| [71] |
Gunitseva N, Evteeva M, Korzhenkov A, Patrushev M. A new RNA-dependent Cas12g nuclease. Int J Mol Sci, 2023, 24(23): 17105.
pmid: 38069429 |
| [72] |
Li Z, Zhang H, Xiao RJ, Han RJ, Chang LF. Cryo-EM structure of the RNA-guided ribonuclease Cas12g. Nat Chem Biol, 2021, 17(4): 387-393.
pmid: 33495647 |
| [73] |
Yang J, Li XZC, He QQ, Wang XS, Tang JJ, Wang TY, Zhang Y, Yu FY, Zhang SQ, Liu ZK, Zhang LL, Liao FM, Yin H, Zhao HY, Deng ZQ, Zhang H. Structural basis for the activity of the type VII CRISPR-Cas system. Nature, 2024, 633(8029): 465-472.
pmid: 39143216 |
| [74] |
Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT, Kellner MJ, Regev A, Lander ES, Voytas DF, Ting AY, Zhang F. RNA targeting with CRISPR-Cas13. Nature, 2017, 550(7675): 280-284.
pmid: 28976959 |
| [75] |
Aman R, Ali Z, Butt H, Mahas A, Aljedaani F, Khan MZ, Ding SW, Mahfouz M. RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol, 2018, 19(1): 1.
pmid: 29301551 |
| [76] |
Aman R, Mahas A, Butt H, Aljedaani F, Mahfouz M. Engineering RNA virus interference via the CRISPR/ Cas13 machinery in Arabidopsis. Viruses, 2018, 10(12): 732.
pmid: 30572690 |
| [77] |
Freije CA, Myhrvold C, Boehm CK, Lin AE, Welch NL, Carter A, Metsky HC, Luo CY, Abudayyeh OO, Gootenberg JS, Yozwiak NL, Zhang F, Sabeti PC. Programmable inhibition and detection of RNA viruses using Cas13. Mol Cell, 2019, 76(5): 826-837.e11.
pmid: 31607545 |
| [78] |
Abbott TR, Dhamdhere G, Liu YX, Lin XQ, Goudy L, Zeng LP, Chemparathy A, Chmura S, Heaton NS, Debs R, Pande T, Endy D, La Russa MF, Lewis DB, Qi LS. Development of CRISPR as an antiviral strategy to combat SARS-CoV-2 and influenza. Cell, 2020, 181(4): 865-876.e12.
pmid: 32353252 |
| [79] |
Huynh N, Depner N, Larson R, King-Jones K. A versatile toolkit for CRISPR-Cas13-based RNA manipulation in Drosophila. Genome Biol, 2020, 21(1): 279.
pmid: 33203452 |
| [80] |
Cui J, Techakriengkrai N, Nedumpun T, Suradhat S. Abrogation of PRRSV infectivity by CRISPR-Cas13b- mediated viral RNA cleavage in mammalian cells. Sci Rep, 2020, 10(1): 9617.
pmid: 32541822 |
| [81] |
Tng PYL, Carabajal Paladino L, Verkuijl SAN, Purcell J, Merits A, Leftwich PT, Fragkoudis R, Noad R, Alphey L. Cas13b-dependent and Cas13b-independent RNA knockdown of viral sequences in Mosquito cells following guide RNA expression. Commun Biol, 2020, 3(1): 413.
pmid: 32737398 |
| [82] |
Blanchard EL, Vanover D, Bawage SS, Tiwari PM, Rotolo L, Beyersdorf J, Peck HE, Bruno NC, Hincapie R, Michel F, Murray J, Sadhwani H, Vanderheyden B, Finn MG, Brinton MA, Lafontaine ER, Hogan RJ, Zurla C, Santangelo PJ. Treatment of influenza and SARS-CoV-2 infections via mRNA-encoded Cas13a in rodents. Nat Biotechnol, 2021, 39(6): 717-726.
pmid: 33536629 |
| [83] |
Kiga K, Tan XE, Ibarra-Chávez R, Watanabe S, Aiba Y, Sato’o Y, Li FY, Sasahara T, Cui BT, Kawauchi M, Boonsiri T, Thitiananpakorn K, Taki Y, Azam AH, Suzuki M, Penadés JR, Cui LZ. Development of CRISPR-Cas13a-based antimicrobials capable of sequence-specific killing of target bacteria. Nat Commun, 2020, 11: 2934.
pmid: 32523110 |
| [84] |
Meeske AJ, Nakandakari-Higa S, Marraffini LA. Cas13- induced cellular dormancy prevents the rise of CRISPR- resistant bacteriophage. Nature, 2019, 570(7760): 241-245.
pmid: 31142834 |
| [85] |
Cox DBT, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J, Zhang F. RNA editing with CRISPR-Cas13. Science, 2017, 358(6366): 1019-1027.
pmid: 29070703 |
| [86] |
Abudayyeh OO, Gootenberg JS, Franklin B, Koob J, Kellner MJ, Ladha A, Joung J, Kirchgatterer P, Cox DBT, Zhang F. A cytosine deaminase for programmable single-base RNA editing. Science, 2019, 365(6451): 382-386.
pmid: 31296651 |
| [87] |
Kannan S, Altae-Tran H, Jin X, Madigan VJ, Oshiro R, Makarova KS, Koonin EV, Zhang F. Compact RNA editors with small Cas13 proteins. Nat Biotechnol, 2022, 40(2): 194-197.
pmid: 34462587 |
| [88] |
Liu YJ, Mao SS, Huang SS, Li YQ, Chen YX, Di MH, Huang XX, Lv JJ, Wang XX, Ge JY, Shen SX, Zhang XM, Liu DH, Huang XX, Chi T. REPAIRx, a specific yet highly efficient programmable a > I RNA base editor. EMBO J, 2020, 39(22): e104748.
pmid: 33058207 |
| [89] |
Li G, Wang YH, Li XY, Wang YZ, Huang XX, Gao JN, Hu XX. Developing PspCas13b-based enhanced RESCUE system, eRESCUE, with efficient RNA base editing. Cell Commun Signal, 2021, 19(1): 84.
pmid: 34380502 |
| [90] |
Xiao QQ, Xu ZJ, Xue YY, Xu CL, Han L, Liu YH, Wang F, Zhang RZ, Han S, Wang X, Li GL, Li HW, Yang H, Shu YL. Rescue of autosomal dominant hearing loss by in vivo delivery of mini dCas13X-derived RNA base editor. Sci Transl Med, 2022, 14(654): eabn0449.
pmid: 35857824 |
| [91] |
Meyer KD, Jaffrey SR. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat Rev Mol Cell Biol, 2014, 15(5): 313-326.
pmid: 24713629 |
| [92] |
Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol, 2019, 20(10): 608-624.
pmid: 31520073 |
| [93] |
Chang C, Ma G, Cheung E, Hutchins AP. A programmable system to methylate and demethylate N6-methyladenosine (m6A) on specific RNA transcripts in mammalian cells. J Biol Chem, 2022, 298(11): 102525.
pmid: 36162509 |
| [94] |
Mo J, Chen ZG, Qin SS, Li S, Liu CG, Zhang L, Ran RX, Kong Y, Wang F, Liu SM, Zhou Y, Zhang XL, Weng XC, Zhou X. TRADES: targeted RNA demethylation by SunTag system. Adv Sci (Weinh), 2020, 7(19): 2001402.
pmid: 33042753 |
| [95] |
Liu XM, Zhou J, Mao YH, Ji QQ, Qian SB. Programmable RNA N6-methyladenosine editing by CRISPR-Cas9 conjugates. Nat Chem Biol, 2019, 15(9): 865-871.
pmid: 31383972 |
| [96] |
Liu Q, Fang LM, Wu CJ. Alternative splicing and isoforms: from mechanisms to diseases. Genes (Basel), 2022, 13(3): 401.
pmid: 35327956 |
| [97] |
Du MH, Jillette N, Zhu JJ, Li S, Cheng AW. CRISPR artificial splicing factors. Nat Commun, 2020, 11(1): 2973.
pmid: 32532987 |
| [98] |
Fiflis DN, Rey NA, Venugopal-Lavanya H, Sewell B, Mitchell-Dick A, Clements KN, Milo S, Benkert AR, Rosales A, Fergione S, Asokan A. Repurposing CRISPR-Cas13 systems for robust mRNA trans-splicing. Nat Commun, 2024, 15(1): 2325.
pmid: 38485709 |
| [99] |
Cai R, Lv R, Shi X, Yang G, Jin J. CRISPR/dCas9 tools: Epigenetic mechanism and application in gene transcriptional regulation. Int J Mol Sci, 2023, 24(19): 14865.
pmid: 37834313 |
| [100] | Montagud-Martínez R, Márquez-Costa R, Rodrigo G. Programmable regulation of translation by harnessing the CRISPR-Cas13 system. Chem Commun (Camb), 2023, 59(18): 2616-2619. |
| [101] |
Apostolopoulos A, Kawamoto N, Chow SYA, Tsuiji H, Ikeuchi Y, Shichino Y, Iwasaki S. dCas13-mediated translational repression for accurate gene silencing in mammalian cells. Nat Commun, 2024, 15(1): 2205.
pmid: 38467613 |
| [102] |
Otoupal PB, Cress BF, Doudna JA, Schoeniger JS. CRISPR-RNAa: targeted activation of translation using dCas13 fusions to translation initiation factors. Nucleic Acids Res, 2022, 50(15): 8986-8998.
pmid: 35950485 |
| [103] |
Cao CC, Li AL, Xu CJ, Wu BR, Liu J, Liu YC. Enhancement of protein translation by CRISPR/ dCasRx coupled with SINEB2 repeat of noncoding RNAs. Nucleic Acids Res, 2023, 51(6): e33.
pmid: 36715335 |
| [104] |
East-Seletsky A, O’Connell MR, Knight SC, Burstein D, Cate JHD, Tjian R, Doudna JA. Two distinct RNase activities of CRISPR-C2c2 enable guide RNA processing and RNA detection. Nature, 2016, 538(7624): 270-273.
pmid: 27669025 |
| [105] |
Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, Verdine V, Donghia N, Daringer NM, Freije CA, Myhrvold C, Bhattacharyya RP, Livny J, Regev A, Koonin EV, Hung DT, Sabeti PC, Collins JJ, Zhang F. Nucleic acid detection with CRISPR-Cas13a/ C2c2. Science, 2017, 356(6336): 438-442.
pmid: 28408723 |
| [106] |
Gootenberg JS, Abudayyeh OO, Kellner MJ, Joung J, Collins JJ, Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science, 2018, 360(6387): 439-444.
pmid: 29449508 |
| [107] |
Shan YY, Zhou XM, Huang R, Xing D. High-fidelity and rapid quantification of miRNA combining crRNA programmability and CRISPR/Cas13a trans-cleavage activity. Anal Chem, 2019, 91(8): 5278-5285.
pmid: 30873832 |
| [108] |
Qin PW, Park M, Alfson KJ, Tamhankar M, Carrion R, Patterson JL, Griffiths A, He Q, Yildiz A, Mathies R, Du K. Rapid and fully microfluidic ebola virus detection with CRISPR-Cas13a. ACS Sens, 2019, 4(4): 1048-1054.
pmid: 30860365 |
| [109] |
Tian T, Shu BW, Jiang YZ, Ye MM, Liu L, Guo ZH, Han ZP, Wang Z, Zhou XM. An ultralocalized Cas13a assay enables universal and nucleic acid amplification-free single-molecule RNA diagnostics. ACS Nano, 2021, 15(1): 1167-1178.
pmid: 33498106 |
| [110] |
Santiago-Frangos A, Hall LN, Nemudraia A, Nemudryi A, Krishna P, Wiegand T, Wilkinson RA, Snyder DT, Hedges JF, Cicha C, Lee HH, Graham A, Jutila MA, Taylor MP, Wiedenheft B. Intrinsic signal amplification by type III CRISPR-Cas systems provides a sequence-specific SARS-CoV-2 diagnostic. Cell Rep Med, 2021, 2(6): 100319.
pmid: 34075364 |
| [111] |
Nemudraia A, Nemudryi A, Buyukyoruk M, Scherffius AM, Zahl T, Wiegand T, Pandey S, Nichols JE, Hall LN, McVey A, Lee HH, Wilkinson RA, Snyder LR, Jones JD, Koutmou KS, Santiago-Frangos A, Wiedenheft B. Sequence-specific capture and concentration of viral RNA by type III CRISPR system enhances diagnostic. Nat Commun, 2022, 13(1): 7762.
pmid: 36522348 |
| [112] |
Yang LZ, Wang Y, Li SQ, Yao RW, Luan PF, Wu H, Carmichael GG, Chen LL. Dynamic imaging of RNA in living cells by CRISPR-Cas13 systems. Mol Cell, 2019, 76(6): 981-997.e7.
pmid: 31757757 |
| [113] |
Wang HF, Nakamura M, Abbott TR, Zhao DH, Luo KW, Yu C, Nguyen CM, Lo A, Daley TP, La Russa M, Liu YX, Qi LS. CRISPR-mediated live imaging of genome editing and transcription. Science, 2019, 365(6459): 1301-1305.
pmid: 31488703 |
| [114] |
Davis BJ, O’Connell MR. Put on your para-spectacles: the development of optimized CRISPR-Cas13-based approaches to image RNA dynamics in real time. Mol Cell, 2020, 77(2): 207-209.
pmid: 31951545 |
| [115] | Xia CL, Colognori D, Jiang XY, Xu K, Doudna JA. Single-molecule live-cell RNA imaging with CRISPR- Csm. Nat Biotechnol, 2025, doi: 10.1038/s41587-024-02540-5. |
| [116] |
Sun NH, Chen DY, Ye LP, Sheng G, Gong JJ, Chen BH, Lu YM, Han F. CRISPR-Sunspot: imaging of endogenous low-abundance RNA at the single-molecule level in live cells. Theranostics, 2020, 10(24): 10993-11012.
pmid: 33042266 |
| [117] |
Zhang ZH, Sun WP, Shi TZ, Lu PF, Zhuang M, Liu JL. Capturing RNA-protein interaction via CRUIS. Nucleic Acids Res, 2020, 48(9): e52.
pmid: 32140725 |
| [118] |
Han S, Zhao BS, Myers SA, Carr SA, He C, Ting AY. RNA-protein interaction mapping via MS2- or Cas13-based APEX targeting. Proc Natl Acad Sci USA, 2020, 117(36): 22068-22079.
pmid: 32839320 |
| [119] |
Chen BW, Shi HY, Zhang J, Zhou C, Han MM, Jiang WX, Lai YT, Tu X, Li HB. CRISPR-based RNA-binding protein mapping in live cells. Biochem Biophys Res Commun, 2021, 583: 79-85.
pmid: 34735883 |
| [120] |
Grünewald J, Zhou RH, Garcia SP, Iyer S, Lareau CA, Aryee MJ, Joung JK. Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature, 2019, 569(7756): 433-437.
pmid: 30995674 |
| [121] |
Mao SS, Liu YJ, Huang SS, Huang XX, Chi T. Site-directed RNA editing (SDRE): Off-target effects and their countermeasures. J Genet Genomics, 2019, 46(11): 531-535.
pmid: 31889638 |
| [122] |
Smargon AA, Shi YJ, Yeo GW. RNA-targeting CRISPR systems from metagenomic discovery to transcriptomic engineering. Nat Cell Biol, 2020, 22(2): 143-150.
pmid: 32015437 |
| [1] | 郭梦玮, 樊友宏, 任国栋. 植物miRNA稳定性与降解的分子基础[J]. 遗传, 2025, 47(8): 944-957. |
| [2] | 师愉倩, 马剑峰, 陈思宇, 周立新, 薛佳, 沈林園, 朱砺, 甘麦邻. 小非编码RNA在雄性生殖发育和代际遗传中研究进展[J]. 遗传, 2025, 47(8): 861-875. |
| [3] | 冯莹, 何晓丽, 刘宇, 王进. 基于质谱的RNA及其修饰分析[J]. 遗传, 2025, 47(8): 885-902. |
| [4] | 杨怀昊, 郑丙莲. 植物小RNA的产生、作用方式、功能及在农业中的应用前景[J]. 遗传, 2025, 47(8): 928-943. |
| [5] | 徐德钰, 周溪, 任玉洁. 基于RNAi的抗病毒免疫[J]. 遗传, 2025, 47(8): 876-884. |
| [6] | 张潇, 于燕, 宁勇, 洪绮雯, 史怀平. MicroRNA促进基因表达研究进展[J]. 遗传, 2025, 47(7): 729-741. |
| [7] | 周泰增, 陈秋阳, 杨祎挺, 甘麦邻, 朱砺, 沈林園. MicroRNA对肌肉再生功能调控研究进展[J]. 遗传, 2025, 47(5): 513-532. |
| [8] | 何锐, 郑秀娟, 王宁宁, 李旭颖, 李明其, 辇士晶, 王克维. 基于生物信息学识别肝细胞癌中的PANoptosis相关lncRNAs并构建预后模型[J]. 遗传, 2025, 47(4): 456-475. |
| [9] | 倪嘉欣, 张蔚. 蝶翅花纹的演化发育生物学研究进展[J]. 遗传, 2025, 47(2): 258-270. |
| [10] | 潘东霞, 王辉, 熊本海, 唐湘方. CRISPR-Cas基因编辑技术在羊生产应用中研究进展[J]. 遗传, 2024, 46(9): 690-700. |
| [11] | 杨森, 马宝霞, 钱泓润, 崔婕妤, 张潇筠, 李利达, 魏泽辉, 张智英, 王建刚, 徐坤. 基于小型化Cas蛋白的CRISPR/Gal4BD-Cas供体适配基因编辑系统研究[J]. 遗传, 2024, 46(9): 716-726. |
| [12] | 吴岳阳, 周小燕, 吴玉峰, 黄驹. NMD途径功能缺陷对水稻表型及转录组的影响[J]. 遗传, 2024, 46(7): 540-551. |
| [13] | 胡玉龙, 杨芳, 陈彦潼, 谌烁楷, 闫煜博, 张跃博, 吴晓林, 汪加明, 何俊, 高宁. 整合mRNA转录本与基因组信息的基因组选择方法研究[J]. 遗传, 2024, 46(7): 560-569. |
| [14] | 马宝霞, 杨森, 吕明, 王昱人, 常立业, 韩艺帆, 王建刚, 郭杨, 徐坤. 不同CRISPR/Cas9供体适配基因编辑系统的比较及优化研究[J]. 遗传, 2024, 46(6): 466-477. |
| [15] | 张译文, 黄琴, 吴艳芸, 孙月, 韦永龙. LIN28A/B在肿瘤发生发展中的作用研究进展[J]. 遗传, 2024, 46(6): 452-465. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||

www.chinagene.cn
备案号:京ICP备09063187号-4
总访问:,今日访问:,当前在线:
