Hereditas(Beijing) ›› 2023, Vol. 45 ›› Issue (12): 1114-1127.doi: 10.16288/j.yczz.23-233
• Review • Previous Articles Next Articles
Application and prospect of gene chip in genetic breeding of livestock and poultry
Jiahao Wang1( ), Qingyao Zhao1, Yueling Zhou1, Liangyu Shi2, Chuduan Wang1(
), Qingyao Zhao1, Yueling Zhou1, Liangyu Shi2, Chuduan Wang1( ), Ying Yu1(
), Ying Yu1( )
)
- 1. College of Animal Science and Technology, China Agricultural University, Beijing 100193, China
2. School of Animal Science and Nutritional Engineering, Wuhan Polytechnic University, Wuhan 430023, China
-
Received:2023-09-06Revised:2023-10-05Online:2023-12-20Published:2023-11-08 -
Contact:Chuduan Wang,Ying Yu E-mail:2423280404@qq.com;cdwang@cau.edu.cn;yuying@cau.edu.cn -
Supported by:National Key R&D Program of China(2021YFD1200903);National Key R&D Program of China(2021YFD1200900);National Dairy Industry Technology System Project(CARS-36);National Natural Science Foundation of China-Pakistan Science Foundation Join Project(31961143009)
Cite this article
Jiahao Wang, Qingyao Zhao, Yueling Zhou, Liangyu Shi, Chuduan Wang, Ying Yu. Application and prospect of gene chip in genetic breeding of livestock and poultry[J]. Hereditas(Beijing), 2023, 45(12): 1114-1127.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
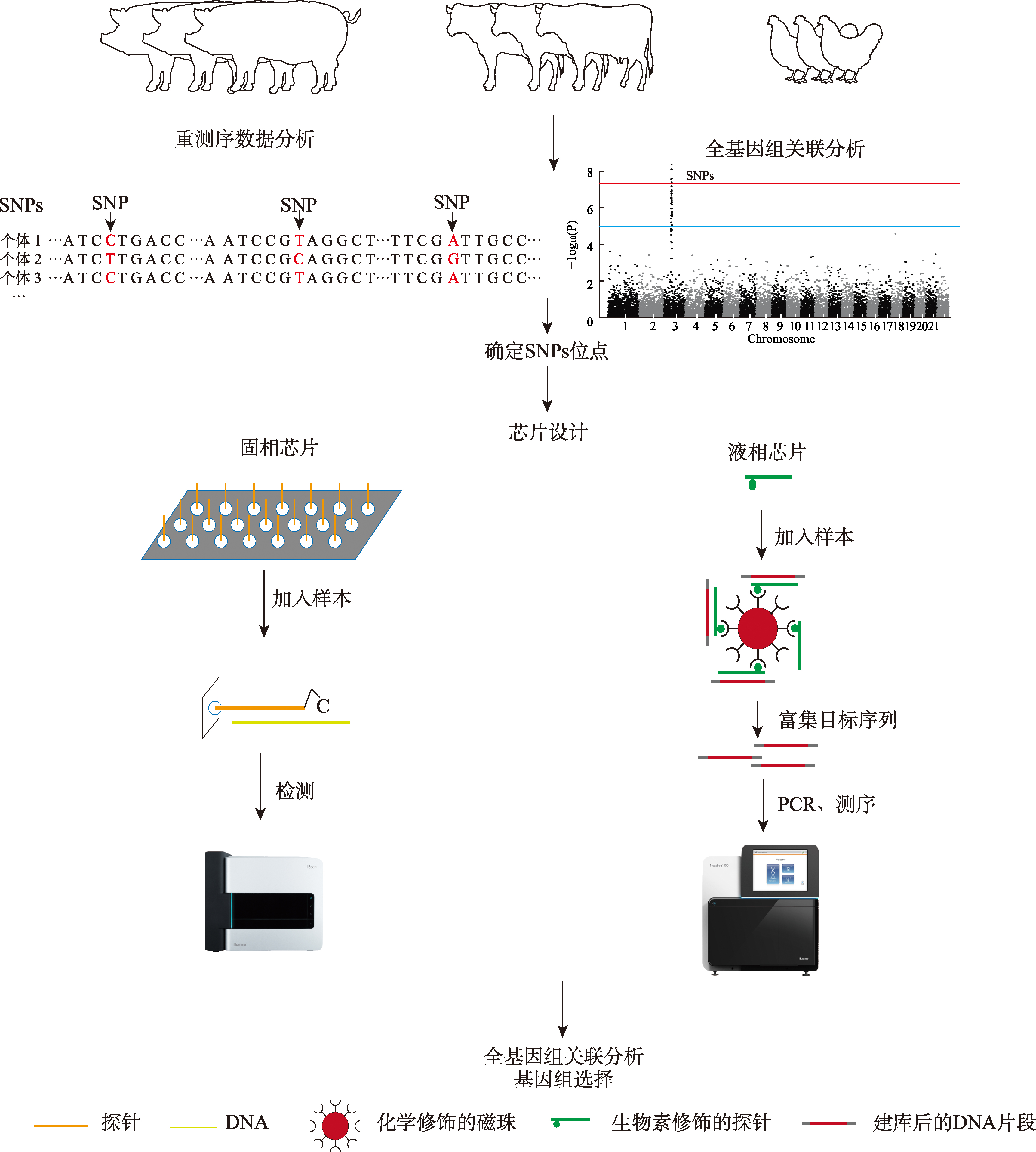
Table 1
SNP chips commonly used on livestock and poultry"
| 物种 | 芯片名称 | 类型 | 开发单位 | 文献 |
|---|---|---|---|---|
| 牛 | BovineSNP50 Genotyping BeadChip | 固相芯片 | 美国Illumina公司 | [ |
| CCSC-I(奶牛乳房健康分子检测芯片-I) | 固相芯片 | 中国农业大学 | [ | |
| BovineHD Genotyping BeadChip | 固相芯片 | 美国Illumina公司 | [ | |
| Axiom Genome-Wide BOS 1 Array | 固相芯片 | 美国Affymetrix公司 | [ | |
| Bovine3K Genotyping BeadChip | 固相芯片 | 美国Illumina公司 | [ | |
| BovineLD Genotyping BeadChip | 固相芯片 | 美国Illumina公司 | [ | |
| A2奶牛基因检测液相芯片 | 液相芯片 | 山东省农业科学院 | ||
| 元牛一号 | 液相芯片 | 内蒙古元牛繁育科技有限公司 | ||
| 吉牛一号 | 液相芯片 | 吉林省农业科学院 | ||
| 奶牛126K液相基因组育种芯片 | 液相芯片 | 中国农业大学 | ||
| 家养水牛100K高密度液相芯片 | 液相芯片 | 中国农业大学 | ||
| 猪 | PorcineSNP60 Genotyping BeadChip | 固相芯片 | 美国Illumina公司 | [ |
| GenoBaits Porcine SNP50K | 液相芯片 | 中国农业大学 | [ | |
| Genomic Profiler 10k BeadChip | 固相芯片 | 美国Neogen公司 | [ | |
| GGP Porcine 50K | 固相芯片 | 美国Neogen公司 | ||
| GGP Porcine 80K | 固相芯片 | 美国Neogen公司 | ||
| GenoBaits? Porcine 1K Panel | 液相芯片 | 中国农业大学、山东农业大学 | ||
| “Zhongxin-I”Porcine Chip | 固相芯片 | 江西农业大学 | ||
| 羊 | OvineSNP50 BeadChip | 固相芯片 | 美国Illumina公司 | [ |
| 绵羊40K液相芯片 | 液相芯片 | 西北农林科技大学 | [ | |
| 绒山羊66K SNP | 液相芯片 | 内蒙古农业大学 | [ | |
| GoatSNPS0 BeadChip array | 固相芯片 | 美国Illumina公司 | [ | |
| Ovine HD SNP BeadChip | 固相芯片 | 国际绵羊基因组学联盟(ISGC) | [ | |
| 鸡 | 京芯一号 | 固相芯片 | 中国农业科学院北京畜牧兽医研究所 | [ |
| 60K SNP | 固相芯片 | 荷兰瓦赫宁根大学 | [ | |
| 600K SNP | 固相芯片 | 英国安伟捷公司、英国罗斯林研究所 | [ | |
| 凤芯壹号 | 固相芯片 | 中国农业大学 | [ | |
| 酉芯一号 | 液相芯片 | 江苏省家禽科学研究所 | ||
| 神农一号 | 液相芯片 | 河南农业大学 |
| [1] | Tan C, Bian C, Yang D, Li N, Wu ZF, Hu XX. Application of genomic selection in farm animal breeding. Hereditas (Beijing), 2017, 39(11): 1033-1045. |
| 谈成, 边成, 杨达, 李宁, 吴珍芳, 胡晓湘. 基因组选择技术在农业动物育种中的应用. 遗传, 2017, 39(11): 1033-1045. | |
| [2] | Yin LL, Ma YL, Xiang T, Zhu MJ, Yu M, Li XY, Liu XL, Zhao SH. The progress and prospect of genomic selection models. Acta Vet Zootech Sin, 2019, 50(2): 233-242. |
| 尹立林, 马云龙, 项韬, 朱猛进, 余梅, 李新云, 刘小磊, 赵书红. 全基因组选择模型研究进展及展望. 畜牧兽医学报, 2019, 50(2): 233-242. | |
| [3] | Wu CX. Animal genetics. Beijing: Higher Education Press, 2009. |
| 吴常信. 动物遗传学. 北京: 高等教育出版社, 2009. | |
| [4] | Zhang Y. Animal breeding. Beijing: China Agriculture Press, 2001. |
| 张沅. 家畜育种学. 北京: 中国农业出版社, 2001. | |
| [5] | Falconer DS, Mackay TFC. Introduction to quantitative genetics. Harlow, UK: Longman Group Limited, 1996. |
| [6] |
Elmhiri G, Gloaguen C, Grison S, Kereselidze D, Elie C, Tack K, Benderitter M, Lestaevel P, Legendre A, Souidi M. DNA methylation and potential multigenerational epigenetic effects linked to uranium chronic low-dose exposure in gonads of males and females rats. Toxicol Lett, 2018, 282: 64-70.
doi: S0378-4274(17)31381-4 pmid: 29024790 |
| [7] |
Guo XJ, Chen XS, Wang J, Liu ZY, Gaile D, Wu HM, Yu G, Mao GY, Yang ZP, Di Z, Guo XQ, Cao L, Chang PY, Kang BX, Chen JY, Gao W, Ren XF. Multi-generational impacts of arsenic exposure on genome-wide DNA methylation and the implications for arsenic-induced skin lesions. Environ Int, 2018, 119: 250-263.
doi: S0160-4120(18)30152-1 pmid: 29982128 |
| [8] |
Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, Slagboom PE, Lumey LH. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci USA, 2008, 105(44): 17046-17049.
doi: 10.1073/pnas.0806560105 pmid: 18955703 |
| [9] |
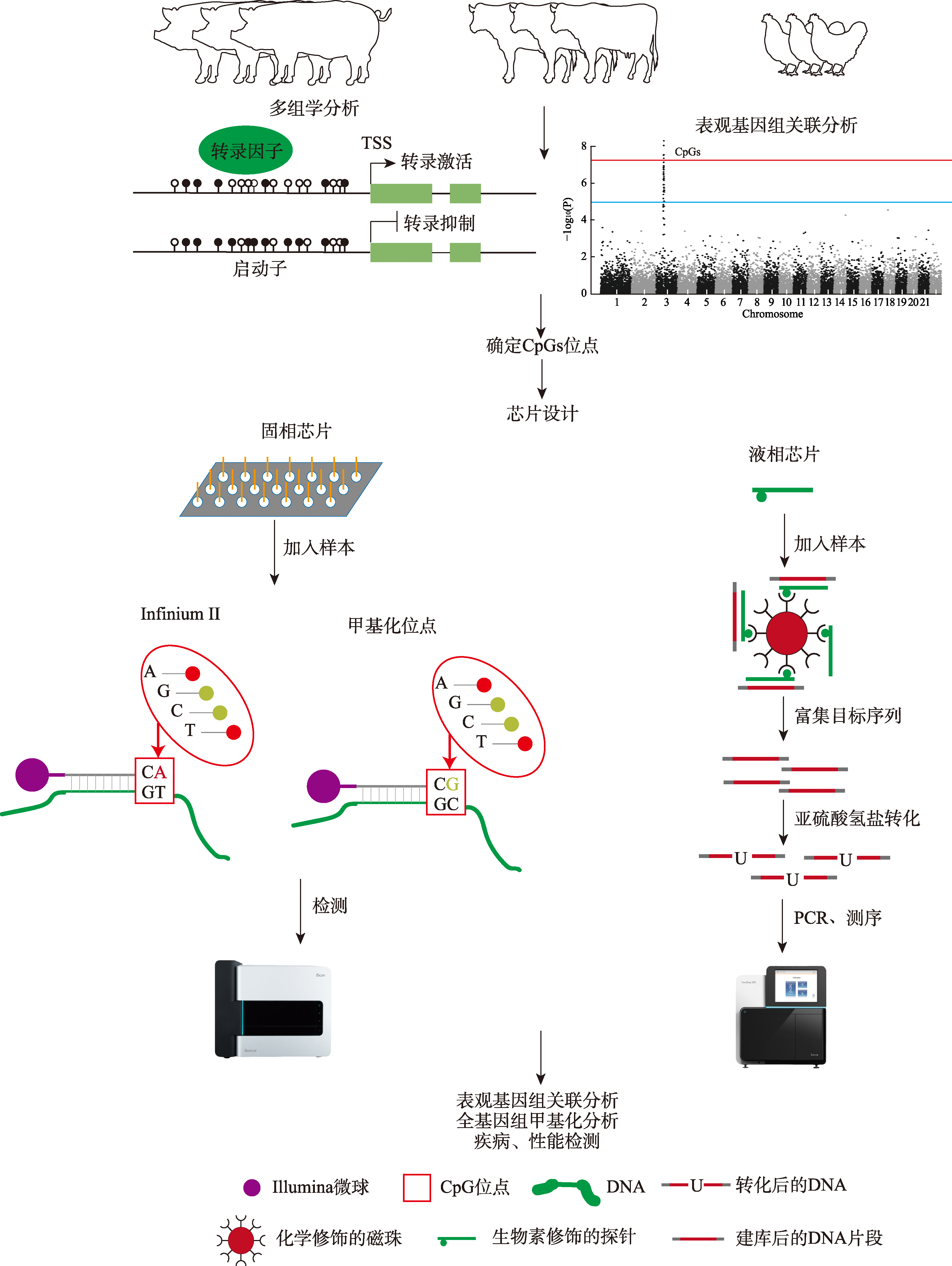
Liu L, Yang N, Xu GY, Liu SL, Wang D, Song JZ, Duan ZY, Yang S, Yu Y. Transgenerational transmission of maternal stimulatory experience in domesticated birds. FASEB J, 2018, 32(12): 7002-7017.
doi: 10.1096/fsb2.v32.12 |
| [10] |
Sakai K, Hara K, Tanemura K. Testicular histone hyperacetylation in mice by valproic acid administration affects the next generation by changes in sperm DNA methylation. PLoS One, 2023, 18(3): e0282898.
doi: 10.1371/journal.pone.0282898 |
| [11] |
Takahashi Y, Morales Valencia M, Yu Y, Ouchi Y, Takahashi K, Shokhirev MN, Lande K, Williams AE, Fresia C, Kurita M, Hishida T, Shojima K, Hatanaka F, Nuñez-Delicado E, Esteban CR, Izpisua Belmonte JC. Transgenerational inheritance of acquired epigenetic signatures at CpG islands in mice. Cell, 2023, 186(4): 715-731. e719.
doi: 10.1016/j.cell.2022.12.047 pmid: 36754048 |
| [12] |
Banta JA, Richards CL. Quantitative epigenetics and evolution. Heredity (Edinb), 2018, 121(3): 210-224.
doi: 10.1038/s41437-018-0114-x |
| [13] |
Fodor SP, Read JL, Pirrung MC, Stryer L, Lu AT, Solas D. Light-directed, spatially addressable parallel chemical synthesis. Science, 1991, 251(4995): 767-773.
doi: 10.1126/science.1990438 pmid: 1990438 |
| [14] |
Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science, 1995, 270(5235): 467-470.
doi: 10.1126/science.270.5235.467 pmid: 7569999 |
| [15] |
McDade RL, Spain MD. Rapid, economical testing in the clinical laboratory: a new flow cytometry-based multiplex system. Clin Immunol Newslett, 1997, 17(10): 154-158.
doi: 10.1016/S0197-1859(00)80021-7 |
| [16] |
Yeakley JM, Fan JB, Doucet D, Luo L, Wickham E, Ye Z, Chee MS, Fu XD. Profiling alternative splicing on fiber-optic arrays. Nat Biotechnol, 2002, 20(4): 353-358.
doi: 10.1038/nbt0402-353 pmid: 11923840 |
| [17] | Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, Fennell T, Giannoukos G, Fisher S, Russ C, Gabriel S, Jaffe DB, Lander ES, Nusbaum C. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol, 2009, 27(2): 182-189. |
| [18] |
Jordan SA, Humphries P.Single nucleotide polymerphism in exon 2 of the BCP gene on 7q31-q35. Hum Mol Genet, 1994, 3(10): 1915-1915.
pmid: 7849733 |
| [19] |
Lander ES. The new genomics: global views of biology. Science, 1996, 274(5287): 536-539.
doi: 10.1126/science.274.5287.536 pmid: 8928008 |
| [20] | 霍艳军, 陈宠霞, 金一. SNPs标记技术及其在牛遗传育种中的应用. 现代畜牧兽医, 2006, (6): 57-60. |
| [21] |
Zou YP, Ge S. A novel molecular marker—SNPs and its application. Biodiversity Sci, 2003, 11(5): 370-382.
doi: 10.17520/biods.2003045 |
|
邹喻苹, 葛颂. 新一代分子标记——SNPs及其应用. 生物多样性, 2003, 11(5): 370-382.
doi: 10.17520/biods.2003045 |
|
| [22] |
Delahunty C, Ankener W, Deng Q, Eng J, Nickerson DA. Testing the feasibility of DNA typing for human identification by PCR and an oligonucleotide ligation assay. Am J Hum Genet, 1996, 58(6): 1239-1246.
pmid: 8651301 |
| [23] |
Fan JB, Chen X, Halushka MK, Berno A, Huang X, Ryder T, Lipshutz RJ, Lockhart DJ, Chakravarti A. Parallel genotyping of human SNPs using generic high-density oligonucleotide tag arrays. Genome Res, 2000, 10(6): 853-860.
pmid: 10854416 |
| [24] |
Hirschhorn JN, Sklar P, Lindblad-Toh K, Lim YM, Ruiz-Gutierrez M, Bolk S, Langhorst B, Schaffner S, Winchester E, Lander ES. SBE-TAGS: an array-based method for efficient single-nucleotide polymorphism genotyping. Proc Natl Acad Sci USA, 2000, 97(22): 12164-12169.
doi: 10.1073/pnas.210394597 pmid: 11035790 |
| [25] |
Fan JB, Oliphant A, Shen R, Kermani BG, Garcia F, Gunderson KL, Hansen M, Steemers F, Butler SL, Deloukas P, Galver L, Hunt S, McBride C, Bibikova M, Rubano T, Chen J, Wickham E, Doucet D, Chang W, Campbell D, Zhang B, Kruglyak S, Bentley D, Haas J, Rigault P, Zhou L, Stuelpnagel J, Chee MS. Highly parallel SNP genotyping. Cold Spring Harb Symp Quant Biol, 2003, 68: 69-78.
pmid: 15338605 |
| [26] | Oliphant A, Barker DL, Stuelpnagel JR, Chee MS. BeadArray technology: enabling an accurate, cost- effective approach to high-throughput genotyping. BioTechniques, 2002, Suppl: 56-58, 60-61. |
| [27] |
Xu YB, Yang QN, Zhen HJ, Xu YF, Sang ZQ, Guo ZF, Peng H, Zhang C, Lan HF, Wang YB, Wu KS, Tao JJ, Zhang JN. Genotyping by target sequencing (GBTS) and its applications. Sci Agric Sin, 2020, 53(15): 2983-3004.
doi: 10.3864/j.issn.0578-1752.2020.15.001 |
|
徐云碧, 杨泉女, 郑洪建, 许彦芬, 桑志勤, 郭子锋, 彭海, 张丛, 蓝昊发, 王蕴波, 吴坤生, 陶家军, 张嘉楠. 靶向测序基因型检测(GBTS)技术及其应用. 中国农业科学, 2020, 53(15): 2983-3004.
doi: 10.3864/j.issn.0578-1752.2020.15.001 |
|
| [28] |
Zhang M, Yang LL, Jia YL, Wang TY. Research progress in the roles of DNA and histone methylations in epigenetic regulation. Biotechnol Bull, 2022, 38(7): 23-30.
doi: 10.13560/j.cnki.biotech.bull.1985.2021-1054 |
|
张淼, 杨露露, 贾岩龙, 王天云. DNA甲基化和组蛋白甲基化修饰的表观遗传调控作用研究进展. 生物技术通报 2022, 38(7): 23-30.
doi: 10.13560/j.cnki.biotech.bull.1985.2021-1054 |
|
| [29] | Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature, 2008, 454(7205): 766-770. |
| [30] |
Jylhävä J, Pedersen NL, Hägg S. Biological age predictors. EBioMedicine, 2017, 21: 29-36.
doi: S2352-3964(17)30142-1 pmid: 28396265 |
| [31] |
Widschwendter M. 5-methylcytosine--the fifth base of DNA: the fifth wheel on a car or a highly promising diagnostic and therapeutic target in cancer? Dis Markers, 2007, 23(1-2): 1-3.
pmid: 17325422 |
| [32] |
Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA, 1992, 89(5): 1827-1831.
doi: 10.1073/pnas.89.5.1827 pmid: 1542678 |
| [33] |
Grunau C, Clark SJ, Rosenthal A. Bisulfite genomic sequencing: systematic investigation of critical experimental parameters. Nucleic Acids Res, 2001, 29(13): E65-e65.
doi: 10.1093/nar/29.13.e65 pmid: 11433041 |
| [34] |
Bibikova M, Lin Z, Zhou L, Chudin E, Garcia EW, Wu B, Doucet D, Thomas NJ, Wang Y, Vollmer E, Goldmann T, Seifart C, Jiang W, Barker DL, Chee MS, Floros J, Fan JB. High-throughput DNA methylation profiling using universal bead arrays. Genome Res, 2006, 16(3): 383-393.
doi: 10.1101/gr.4410706 pmid: 16449502 |
| [35] |
Deng J, Shoemaker R, Xie B, Gore A, LeProust EM, Antosiewicz-Bourget J, Egli D, Maherali N, Park IH, Yu JY, Daley GQ, Eggan K, Hochedlinger K, Thomson J, Wang W, Gao Y, Zhang K. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat Biotechnol, 2009, 27(4): 353-360.
doi: 10.1038/nbt.1530 pmid: 19330000 |
| [36] |
Lee EJ, Pei L, Srivastava G, Joshi T, Kushwaha G, Choi JH, Robertson KD, Wang X, Colbourne JK, Zhang L, Schroth GP, Xu D, Zhang K, Shi H. Targeted bisulfite sequencing by solution hybrid selection and massively parallel sequencing. Nucleic Acids Res, 2011, 39(19): e127.
doi: 10.1093/nar/gkr598 |
| [37] |
Stirzaker C, Taberlay PC, Statham AL, Clark SJ. Mining cancer methylomes: prospects and challenges. Trends Genet, 2014, 30(2): 75-84.
doi: 10.1016/j.tig.2013.11.004 pmid: 24368016 |
| [38] |
Meuwissen TH, Hayes BJ, Goddard ME. Prediction of total genetic value using genome-wide dense marker maps. Genetics, 2001, 157(4): 1819-1829.
doi: 10.1093/genetics/157.4.1819 pmid: 11290733 |
| [39] | 董航言, 黄生强. 全基因组选择在猪育种上的研究进展. 猪业科学, 2015, (4): 104-105. |
| [40] |
Schaeffer LR. Strategy for applying genome-wide selection in dairy cattle. J Anim Breed Genet, 2006, 123(4): 218-223.
doi: 10.1111/j.1439-0388.2006.00595.x pmid: 16882088 |
| [41] |
Matukumalli LK, Lawley CT, Schnabel RD, Taylor JF, Allan MF, Heaton MP, O'Connell J, Moore SS, Smith TP, Sonstegard TS, Van Tassell CP,. Development and characterization of a high density SNP genotyping assay for cattle. PLoS One, 2009, 4(4): e5350.
doi: 10.1371/journal.pone.0005350 |
| [42] |
VanRaden PM, Van Tassell CP, Wiggans GR, Sonstegard TS, Schnabel RD, Taylor JF, Schenkel FS. Invited review: reliability of genomic predictions for North American Holstein bulls. J Dairy Sci, 2009, 92(1): 16-24.
doi: 10.3168/jds.2008-1514 pmid: 19109259 |
| [43] | Pan LN, Qin B. Summary on the development of genomic evaluation system in the United States. China Dairy Cattle, 2015, 307(21): 9-14. |
| 潘丽娜, 秦波. 美国基因组评估系统的发展概述. 中国奶牛, 2015, 307(21): 9-14. | |
| [44] |
Hutchison JL, Cole JB, Bickhart DM. Short communication: use of young bulls in the United States. J Dairy Sci, 2014, 97(5): 3213-3220.
doi: 10.3168/jds.2013-7525 pmid: 24612804 |
| [45] |
Panigrahi M, Kumar H, Saravanan KA, Rajawat D, Sonejita Nayak S, Ghildiyal K, Kaisa K, Parida S, Bhushan B, Dutt T. Trajectory of livestock genomics in South Asia: a comprehensive review. Gene, 2022, 843: 146808.
doi: 10.1016/j.gene.2022.146808 |
| [46] |
Ramos AM, Crooijmans RPMA, Affara NA, Amaral AJ, Archibald AL, Beever JE, Bendixen C, Churcher C, Clark R, Dehais P, Hansen MS, Hedegaard J, Hu ZL, Kerstens HH, Law AS, Megens HJ, Milan D, Nonneman DJ, Rohrer GA, Rothschild MF, Smith TPL, Schnabel RD, Van Tassell CP, Taylor JF, Wiedmann RT, Schook LB, Groenen MAM. Design of a high density SNP genotyping assay in the pig using SNPs identified and characterized by next generation sequencing technology. PLoS One, 2009, 4(8): e6524.
doi: 10.1371/journal.pone.0006524 |
| [47] | 王青来, 李亚兰. 全国首例全基因组选育特级种猪在粤诞生. 农业知识(科学养殖), 2014, (2): 25-25. |
| [48] |
Ibáñez-Escriche N, Forni S, Noguera JL, Varona L. Genomic information in pig breeding: science meets industry needs. Livest Sci, 2014, 166: 94-100.
doi: 10.1016/j.livsci.2014.05.020 |
| [49] |
Kijas JW, Townley D, Dalrymple BP, Heaton MP, Maddox JF, McGrath A, Wilson P, Ingersoll RG, McCulloch R, McWilliam S, Tang D, McEwan J, Cockett N, Oddy VH, Nicholas FW, Raadsma H, International Sheep Genomics Consortium. A genome wide survey of SNP variation reveals the genetic structure of sheep breeds. PLoS One, 2009, 4(3): e4668.
doi: 10.1371/journal.pone.0004668 |
| [50] | Liu RR, Zhao GP, Wen J. Development of genome-wide SNP genotyping arrays for chicken breeding and conservation. China Poultry, 2018, 40(15): 1-6. |
| 刘冉冉, 赵桂苹, 文杰. 鸡基因组育种和保种用SNP芯片研发及应用. 中国家禽, 2018, 40(15): 1-6. | |
| [51] |
Liu RR, Xing SY, Wang J, Zheng MQ, Cui HX, Crooijmans RPMA, Li QH, Zhao GP, Wen J. A new chicken 55K SNP genotyping array. BMC Genomics, 2019, 20(1): 410.
doi: 10.1186/s12864-019-5736-8 pmid: 31117951 |
| [52] | 邱奥, 张梓鹏, 王雪, 罗文学, 王贵江, 丁向东. 猪50K液相芯片基因组选择效果分析. 中国畜牧杂志, 2022, 58(8): 82-86. |
| [53] | Zhang ZP, Xing SY, Qiu A, Zhang N, Wang WW, Qian CS, Zhang JN, Wang CD, Zhang Q, Ding XD.The development of a porcine 50K SNP panel using genotyping by target sequencing and its application1. J Integr Agr, 2023. |
| [54] | Xu L.The design of low-density SNP array for genomic selection in simmental beef cattle[Dissertation]. Chinese Academy of Agricultural Sciences, 2019. |
| 徐玲.肉用西门塔尔牛全基因组选择的低密度SNP芯片设计研究[学位论文]. 中国农业科学院, 2019. | |
| [55] |
Khan MZ, Dari G, Khan A, Yu Y. Genetic polymorphisms of TRAPPC9 and CD4 genes and their association with milk production and mastitis resistance phenotypic traits in Chinese Holstein. Front Vet Sci, 2022, 9: 1008497.
doi: 10.3389/fvets.2022.1008497 |
| [56] |
Khan MZ, Wang D, Liu L, Usman T, Wen H, Zhang R, Liu S, Shi L, Mi S, Xiao W, Yu Y. Significant genetic effects of JAK2 and DGAT1 mutations on milk fat content and mastitis resistance in Holsteins. J Dairy Res, 2019, 86(4): 388-393.
doi: 10.1017/S0022029919000682 |
| [57] | Guo YW.Development and application of 40K liquid gene chip for sheep[Dissertation]. Northwest A&F University, 2022. |
| 郭应威.绵羊40K液相基因芯片的开发与应用[学位论文]. 西北农林科技大学, 2022. | |
| [58] |
Qiao X, Su R, Wang Y, Wang RJ, Yang T, Li XK, Chen W, He SY, Jiang Y, Xu QW, Wan WT, Zhang YL, Zhang WG, Chen J, Liu B, Liu X, Fan YX, Chen DY, Jiang HZ, Fang DM, Liu ZH, Wang XW, Zhang YJ, Mao DQ, Wang ZY, Di R, Zhao QJ, Zhong T, Yang HM, Wang J, Wang W, Dong Y, Chen XL, Xu X, Li JQ. Genome-wide target enrichment-aided chip design: a 66 K SNP chip for Cashmere goat. Sci Rep, 2017, 7(1): 8621.
doi: 10.1038/s41598-017-09285-z |
| [59] |
Rincon G, Weber KL, Van Eenennaam AL, Golden BL, Medrano JF. Hot topic: performance of bovine high- density genotyping platforms in Holsteins and Jerseys. J Dairy Sci, 2011, 94(12): 6116-6121.
doi: 10.3168/jds.2011-4764 pmid: 22118099 |
| [60] |
Wiggans GR, Cooper TA, Vanraden PM, Olson KM, Tooker ME. Use of the Illumina Bovine3K BeadChip in dairy genomic evaluation. J Dairy Sci, 2012, 95(3): 1552-1558.
doi: 10.3168/jds.2011-4985 pmid: 22365235 |
| [61] |
Boichard D, Chung H, Dassonneville R, David X, Eggen A, Fritz S, Gietzen KJ, Hayes BJ, Lawley CT, Sonstegard TS, Van Tassell CP, VanRaden PM, Viaud-Martinez KA, Wiggans GR, Bovine LD Consortium. Design of a bovine low-density SNP array optimized for imputation. PLoS One, 2012, 7(3): e34130.
doi: 10.1371/journal.pone.0034130 |
| [62] |
Derks MFL, Megens HJ, Bosse M, Lopes MS, Harlizius B, Groenen MAM. A systematic survey to identify lethal recessive variation in highly managed pig populations. BMC Genomics, 2017, 18(1): 858.
doi: 10.1186/s12864-017-4278-1 pmid: 29121877 |
| [63] |
Tosser-Klopp G, Bardou P, Bouchez O, Cabau C, Crooijmans R, Dong Y, Donnadieu-Tonon C, Eggen A, Heuven HCM, Jamli S, Jiken AJ, Klopp C, Lawley CT, McEwan J, Martin P, Moreno CR, Mulsant P, Nabihoudine I, Pailhoux E, Palhière I, Rupp R, Sarry J, Sayre BL, Tircazes A, Jun W, Wang W, Zhang WG, International Goat Genome Consortium. Design and characterization of a 52K SNP chip for goats. PLoS One, 2014, 9(1): e86227.
doi: 10.1371/journal.pone.0086227 |
| [64] | Anderson R. Development of a high density (600 K) Illumina ovine SNP chip and its use to fine map the yellow fat locus. In: Plant and Animal Genome XXII Conference. 2014. |
| [65] |
Ma Q, Liu XX, Pan JF, Ma LN, Ma YH, He XH, Zhao QJ, Pu YB, Li YK, Jiang L. Genome-wide detection of copy number variation in Chinese indigenous sheep using an ovine high-density 600 K SNP array. Sci Rep, 2017, 7(1): 912.
doi: 10.1038/s41598-017-00847-9 |
| [66] |
Groenen MAM, Megens HJ, Zare Y, Warren WC, Hillier LW, Crooijmans RPMA, Vereijken A, Okimoto R, Muir WM, Cheng HH. The development and characterization of a 60K SNP chip for chicken. BMC Genomics, 2011, 12(1): 274.
doi: 10.1186/1471-2164-12-274 pmid: 21627800 |
| [67] |
Kranis A, Gheyas AA, Boschiero C, Turner F, Yu L, Smith S, Talbot R, Pirani A, Brew F, Kaiser P, Hocking PM, Fife M, Salmon N, Fulton J, Strom TM, Haberer G, Weigend S, Preisinger R, Gholami M, Qanbari S, Simianer H, Watson KA, Woolliams JA, Burt DW. Development of a high density 600K SNP genotyping array for chicken. BMC Genomics, 2013, 14: 59.
doi: 10.1186/1471-2164-14-59 pmid: 23356797 |
| [68] |
Liu J, Shen QM, Bao HG. Comparison of seven SNP calling pipelines for the next-generation sequencing data of chickens. PLoS One, 2022, 17(1): e0262574.
doi: 10.1371/journal.pone.0262574 |
| [69] |
Mattei AL, Bailly N, Meissner A. DNA methylation: a historical perspective. Trends Genet, 2022, 38(7): 676-707.
doi: 10.1016/j.tig.2022.03.010 pmid: 35504755 |
| [70] |
Liu SL, Chen SQ, Cai WT, Yin HW, Liu AX, Li YH, Liu GE, Wang YC, Yu Y, Zhang SL. Divergence analyses of sperm DNA methylomes between monozygotic twin AI bulls. Epigenomes, 2019, 3(4): 21.
doi: 10.3390/epigenomes3040021 |
| [71] |
Yang YL, Fan XH, Yan JY, Chen MY, Zhu M, Tang YJ, Liu SY, Tang ZL. A comprehensive epigenome atlas reveals DNA methylation regulating skeletal muscle development. Nucleic Acids Res, 2021, 49(3): 1313-1329.
doi: 10.1093/nar/gkaa1203 pmid: 33434283 |
| [72] | Wang XS, Zhao HM, Tang T, Li K, Qian X, Zhang X, Tu Y, Zheng YS, Wang HD, Yu Y. DNA methylation modification of CD4 gene and its application in livestock diseases-resistant breeding. Acta Vet Zootech Sin, 2017, 48(10): 1796-1806. |
| 王晓铄, 赵会敏, 唐天, 李凯, 钱旭, 张雪, 图雅, 郑云胜, 王怀栋, 俞英. CD4基因的DNA甲基化修饰在家畜抗病育种中的应用. 畜牧兽医学报, 2017, 48(10): 1796-1806. | |
| [73] |
Ibeagha-Awemu EM, Yu Y. Consequence of epigenetic processes on animal health and productivity: is additional level of regulation of relevance? Anim Front, 2021, 11(6): 7-18.
doi: 10.1093/af/vfab057 pmid: 34934525 |
| [74] |
Richards EJ. Population epigenetics. Curr Opin Genet Dev, 2008, 18(2): 221-226.
doi: 10.1016/j.gde.2008.01.014 pmid: 18337082 |
| [75] |
Zhao LN, Liu D, Xu J, Wang ZY, Chen Y, Lei CG, Li Y, Liu GY, Jiang YS. The framework for population epigenetic study. Brief Bioinform, 2018, 19(1): 89-100.
doi: 10.1093/bib/bbw098 pmid: 27760738 |
| [76] |
Hu YD, Morota G, Rosa GJM, Gianola D. Prediction of plant height in arabidopsis thaliana using DNA methylation data. Genetics, 2015, 201(2): 779-793.
doi: 10.1534/genetics.115.177204 pmid: 26253546 |
| [77] | Chen SQ.Identification of ruminant epigenomic evolutionary features and their application in Holstein genomic selection[Dissertation]. China Agricultural University, 2022. |
| 陈思倩.反刍动物表观基因组进化特征鉴定及其在荷斯坦牛基因组选择中应用效果的研究[学位论文]. 中国农业大学, 2022. | |
| [78] |
Yang YL, Zhou R, Li K. Future livestock breeding: precision breeding based on multi-omics information and population personalization. J Integr Agr, 2017, 16(12): 2784-2791.
doi: 10.1016/S2095-3119(17)61780-5 |
| [79] |
Christiansen SN, Andersen JD, Kampmann M-L, Liu J, Andersen MM, Tfelt-Hansen J, Morling N. Reproducibility of the Infinium methylationEPIC BeadChip assay using low DNA amounts. Epigenetics, 2022, 17(12): 1636-1645.
doi: 10.1080/15592294.2022.2051861 |
| [80] |
Bibikova M, Le J, Barnes B, Saedinia-Melnyk S, Zhou LX, Shen R, Gunderson KL. Genome-wide DNA methylation profiling using Infinium® assay. Epigenomics, 2009, 1(1): 177-200.
doi: 10.2217/epi.09.14 pmid: 22122642 |
| [81] |
Pidsley R, Zotenko E, Peters TJ, Lawrence MG, Risbridger GP, Molloy P, Van Djik S, Muhlhausler B, Stirzaker C, Clark SJ. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole- genome DNA methylation profiling. Genome Biol, 2016, 17(1): 208.
pmid: 27717381 |
| [82] |
Wan ES, Qiu WL, Baccarelli A, Carey VJ, Bacherman H, Rennard SI, Agusti A, Anderson W, Lomas DA, Demeo DL. Cigarette smoking behaviors and time since quitting are associated with differential DNA methylation across the human genome. Hum Mol Genet, 2012, 21(13): 3073-3082.
doi: 10.1093/hmg/dds135 pmid: 22492999 |
| [83] |
Rakyan VK, Down TA, Balding DJ, Beck S. Epigenome-wide association studies for common human diseases. Nat Rev Genet, 2011, 12(8): 529-541.
doi: 10.1038/nrg3000 pmid: 21747404 |
| [84] |
Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, Delano D, Zhang L, Schroth GP, Gunderson KL, Fan JB, Shen R. High density DNA methylation array with single CpG site resolution. Genomics, 2011, 98(4): 288-295.
doi: 10.1016/j.ygeno.2011.07.007 pmid: 21839163 |
| [85] | Xu J, Zhao LN, Liu D, Hu SM, Song XL, Li J, Lv HC, Duan L, Zhang MM, Jiang QH, Liu GY, Jin SL, Liao MZ, Zhang M, Feng RN, Kong FW, Xu LD, Jiang YS.EWAS: epigenome-wide association study software 2.0. Bioinformatics(Oxford, England), 2018, 34(15): 2657-2658. |
| [86] |
Xiong Z, Yang F, Li MW, Ma YK, Zhao W, Wang GL, Li ZH, Zheng XC, Zou D, Zong WT, Kang HG, Jia YK, Li RJ, Zhang Z, Bao YM. EWAS open platform: integrated data, knowledge and toolkit for epigenome-wide association study. Nucleic Acids Res, 2022, 50(D1): D1004-D1009.
doi: 10.1093/nar/gkab972 |
| [87] |
Wahl S, Drong A, Lehne B, Loh M, Scott WR, Kunze S, Tsai PC, Ried JS, Zhang WH, Yang YW, Tan SL, Fiorito G, Franke L, Guarrera S, Kasela S, Kriebel J, Richmond RC, Adamo M, Afzal U, Ala-Korpela M, Albetti B, Ammerpohl O, Apperley JF, Beekman M, Bertazzi PA, Black SL, Blancher C, Bonder MJ, Brosch M, Carstensen-Kirberg M, de Craen AJM, de Lusignan S, Dehghan A, Elkalaawy M, Fischer K, Franco OH, Gaunt TR, Hampe J, Hashemi M, Isaacs A, Jenkinson A, Jha S, Kato N, Krogh V, Laffan M, Meisinger C, Meitinger T, Mok ZY, Motta V, Ng HK, Nikolakopoulou Z, Nteliopoulos G, Panico S, Pervjakova N, Prokisch H, Rathmann W, Roden M, Rota F, Rozario MA, Sandling JK, Schafmayer C, Schramm K, Siebert R, Slagboom PE, Soininen P, Stolk L, Strauch K, Tai ES, Tarantini L, Thorand B, Tigchelaar EF, Tumino R, Uitterlinden AG, van Duijn C, van Meurs JBJ, Vineis P, Wickremasinghe AR, Wijmenga C, Yang TP, Yuan W, Zhernakova A, Batterham RL, Smith GD, Deloukas P, Heijmans BT, Herder C, Hofman A, Lindgren CM, Milani L, van der Harst P, Peters A, Illig T, Relton CL, Waldenberger M, Järvelin MR, Bollati V, Soong R, Spector TD, Scott J, McCarthy MI, Elliott P, Bell JT, Matullo G, Gieger C, Kooner JS, Grallert H, Chambers JC. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature, 2017, 541(7635): 81-86.
doi: 10.1038/nature20784 |
| [88] |
Li SB, Mancuso N, Metayer C, Ma XM, de Smith AJ, Wiemels JL. Incorporation of DNA methylation quantitative trait loci (mQTLs) in epigenome-wide association analysis: application to birthweight effects in neonatal whole blood. Clin Epigenetics, 2022, 14(1): 158.
doi: 10.1186/s13148-022-01385-6 pmid: 36457128 |
| [89] |
Can SN, Nunn A, Galanti D, Langenberger D, Becker C, Volmer K, Heer K, Opgenoorth L, Fernandez-Pozo N, Rensing SA. The EpiDiverse plant epigenome-wide association studies (EWAS) pipeline. Epigenomes, 2021, 5(2): 12.
doi: 10.3390/epigenomes5020012 |
| [90] |
Ratan P, Rubbi L, Thompson M, Naresh K, Waddell J, Jones B, Pellegrini M. Epigenetic aging in cows is accelerated by milk production. Epigenetics, 2023, 18(1): 2240188.
doi: 10.1080/15592294.2023.2240188 |
| [91] |
Takeda K, Kobayashi E, Ogata K, Imai A, Sato S, Adachi H, Hoshino Y, Nishino K, Inoue M, Kaneda M, Watanabe S. Differentially methylated CpG sites related to fertility in Japanese Black bull spermatozoa: epigenetic biomarker candidates to predict sire conception rate. J Reprod Dev, 2021, 67(2): 99-107.
doi: 10.1262/jrd.2020-137 |
| [92] | Yang XN, Zhao MM, Zhang L. Application process of artificial insemination in animal husbandry. Heilongjiang journal of animal reproduction, 2023, 31(1): 43-49, 60 |
| 杨轩宁, 赵萌萌, 张鲁. 人工授精技术在畜牧生产中的应用历程. 黑龙江动物繁殖, 2023, 31(1): 43-49, 60. | |
| [93] | 寇海军, 幸福. 猪的人工授精技术. 甘肃畜牧兽医, 2013, 43(4): 37-38. |
| [94] |
Knox RV. Artificial insemination in pigs today. Theriogenology, 2016, 85(1): 83-93.
doi: 10.1016/j.theriogenology.2015.07.009 pmid: 26253434 |
| [95] |
Wang X, Li WL, Feng X, Li JB, Liu GE, Fang LZ, Yu Y. Harnessing male germline epigenomics for the genetic improvement in cattle. J Anim Sci Biotechnol, 2023, 14(1): 76.
doi: 10.1186/s40104-023-00874-9 |
| [96] |
Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet, 2001, 2(1): 21-32.
doi: 10.1038/35047554 pmid: 11253064 |
| [97] | Wang DX, Zhao HB. Progress of cattle imprinted genes. China Cattle Science, 2022, 48(6): 80-84. |
| 王丁香, 赵红波. 牛印记基因的研究进展. 中国牛业科学, 2022, 48(6): 80-84. | |
| [98] | Zhao RL, Jia CQ, Li AC, Zhang Y, Su JM. Mechanism of erasure, establishment and maintenance of mammalian genomic imprinting. Chinese J Cell Biology, 2020, 42(3): 512-518. |
| 赵若琳, 贾晨琪, 李艾聪, 张涌, 苏建民. 哺乳动物基因组印记的擦除、建立和维持机理. 中国细胞生物学学报, 2020, 42(3): 512-518. | |
| [99] | Tian JH, Chen YG, Hu JH, Lei AM. Imprinted genes and cell reprogramming. J Anim Sci Vet Med, 2022, 41(3): 30-33, 36. |
| 田佳卉, 陈昱光, 胡建宏, 雷安民. 印记基因与细胞重编程. 畜牧兽医杂志, 2022, 41(3): 30-33, 36. | |
| [100] |
Gigante S, Gouil Q, Lucattini A, Keniry A, Beck T, Tinning M, Gordon L, Woodruff C, Speed TP, Blewitt ME, Ritchie ME. Using long-read sequencing to detect imprinted DNA methylation. Nucleic Acids Res, 2019, 47(8): e46.
doi: 10.1093/nar/gkz107 |
| [101] |
Nishio M, Satoh M. Genomic best linear unbiased prediction method including imprinting effects for genomic evaluation. Genet Sel Evol, 2015, 47(1): 32.
doi: 10.1186/s12711-015-0091-y |
| [102] |
Costes V, Chaulot-Talmon A, Sellem E, Perrier JP, Aubert-Frambourg A, Jouneau L, Pontlevoy C, Hozé C, Fritz S, Boussaha M, Le Danvic C, Sanchez MP, Boichard D, Schibler L, Jammes H, Jaffrézic F, Kiefer H. Predicting male fertility from the sperm methylome: application to 120 bulls with hundreds of artificial insemination records. Clin Epigenetics, 2022, 14(1): 54.
doi: 10.1186/s13148-022-01275-x pmid: 35477426 |
| [103] |
Shojaei Saadi HA, Fournier É, Vigneault C, Blondin P, Bailey J, Robert C. Genome-wide analysis of sperm DNA methylation from monozygotic twin bulls. Reprod Fertil Dev, 2017, 29(4): 838-843.
doi: 10.1071/RD15384 |
| [104] | 丁向东, 王楚端, 张勤. 基于液相芯片的猪基因组选择实施新策略. 中国畜牧杂志, 2022, 58(4): 65-69. |
| [105] |
Spector BL, Harrell L, Sante D, Wyckoff GJ, Willig L. The methylome and cell-free DNA: current applications in medicine and pediatric disease. Pediatr Res, 2023, 94(1): 89-95.
doi: 10.1038/s41390-022-02448-3 pmid: 36646885 |
| [106] | Hu Y, Ni H. Advance in human free circulating DNA. Hereditas(Beijing), 2008, 30(7): 815-820. |
| 胡影, 倪虹. 人体游离循环DNA的研究进展. 遗传, 2008, 30(7): 815-820. | |
| [107] |
He WS, Bishop KS. The potential use of cell-free- circulating-tumor DNA as a biomarker for prostate cancer. Expert Rev Mol Diagn, 2016, 16(8): 839-852.
doi: 10.1080/14737159.2016.1197121 |
| [108] |
Qi T, Pan M, Shi HJ, Wang LY, Bai YF, Ge QY. Cell-free DNA fragmentomics: the novel promising biomarker. Int J Mol Sci, 2023, 24(2): 1503.
doi: 10.3390/ijms24021503 |
| [109] |
Sharma M, Verma RK, Kumar S, Kumar V. Computational challenges in detection of cancer using cell-free DNA methylation. Comput Struct Biotechnol J, 2022, 20: 26-39.
doi: 10.1016/j.csbj.2021.12.001 |
| [110] |
Lehmann-Werman R, Neiman D, Zemmour H, Moss J, Magenheim J, Vaknin-Dembinsky A, Rubertsson S, Nellgård B, Blennow K, Zetterberg H, Spalding K, Haller MJ, Wasserfall CH, Schatz DA, Greenbaum CJ, Dorrell C, Grompe M, Zick A, Hubert A, Maoz M, Fendrich V, Bartsch DK, Golan T, Ben Sasson SA, Zamir G, Razin A, Cedar H, James Shapiro AM, Glaser B, Shemer R, Dor Y. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc Natl Acad Sci USA, 2016, 113(13): E 1826-E1834.
doi: 10.1073/pnas.1519286113 pmid: 26976580 |
| [111] |
Loyfer N, Magenheim J, Peretz A, Cann G, Bredno J, Klochendler A, Fox-Fisher I, Shabi-Porat S, Hecht M, Pelet T, Moss J, Drawshy Z, Amini H, Moradi P, Nagaraju S, Bauman D, Shveiky D, Porat S, Dior U, Rivkin G, Or O, Hirshoren N, Carmon E, Pikarsky A, Khalaileh A, Zamir G, Grinbaum R, Abu Gazala M, Mizrahi I, Shussman N, Korach A, Wald O, Izhar U, Erez E, Yutkin V, Samet Y, Rotnemer Golinkin D, Spalding KL, Druid H, Arner P, Shapiro AMJ, Grompe M, Aravanis A, Venn O, Jamshidi A, Shemer R, Dor Y, Glaser B, Kaplan T. A DNA methylation atlas of normal human cell types. Nature, 2023, 613(7943): 355-364.
doi: 10.1038/s41586-022-05580-6 |
| [112] |
Pös Z, Pös O, Styk J, Mocova A, Strieskova L, Budis J, Kadasi L, Radvanszky J, Szemes T. Technical and methodological aspects of cell-free nucleic acids analyzes. Int J Mol Sci, 2020, 21(22): 8634.
doi: 10.3390/ijms21228634 |
| [113] |
Rapado-González Ó, Rodríguez-Ces AM, López-López R, Suárez-Cunqueiro MM. Liquid biopsies based on cell-free DNA as a potential biomarker in head and neck cancer. Jpn Dent Sci Rev, 2023, 59: 289-302.
doi: 10.1016/j.jdsr.2023.08.004 |
| [1] | Mengxuan Xu, Ming Zhou. Advances of RNA polymerase IV in controlling DNA methylation and development in plants [J]. Hereditas(Beijing), 2022, 44(7): 567-580. |
| [2] | Yongqiang Kong, Jinkai Liu, Jiaqi Gu, Jingyi Xu, Yunuo Zheng, Yiliang Wei, Shaoyuan Wu. Optimization scheme of machine learning model for genetic division between northern Han, southern Han, Korean and Japanese [J]. Hereditas(Beijing), 2022, 44(11): 1028-1043. |
| [3] | Linghong Li, Tong Gou, Aixia Ren, Pengcheng Ding, Wen Lin, Xiangyun Wu, Min Sun, Zhiqiang Gao. Progress on genomics and locus of important agronomic traits in Chenopodium quinoa [J]. Hereditas(Beijing), 2022, 44(11): 1009-1027. |
| [4] | Wang Ya'nan, Tao Xu, Wanpeng Wang, Qingzhu Zhang, Xie Li'nan. Role of epigenetic modifications in the development of crops essential traits [J]. Hereditas(Beijing), 2021, 43(9): 858-879. |
| [5] | Xiangqian Zhang, Nan Li, Xinming Xie. Design and exploration of epigenetic comprehensive experiments [J]. Hereditas(Beijing), 2021, 43(12): 1179-1187. |
| [6] | Xinyue Wang, Liang Li, Qiuhui Duan, Dali Li, Jinlian Chen. Effect of Uhrf1 on intestinal development [J]. Hereditas(Beijing), 2021, 43(1): 84-93. |
| [7] | Ming Liu, Yi Li, Yafang Yang, Yuwen Yan, Fan Liu, Caixia Li, Faming Zeng, Wenting Zhao. Human facial shape related SNP analysis in Han Chinese populations [J]. Hereditas(Beijing), 2020, 42(7): 680-690. |
| [8] | Yang Liu, Changchun Sun, Mi Ma, Ling Wang, Wenting Zhao, Quan Ma, Anquan Ji, Jing Liu, Caixia Li. The ancestry inference of Chinese populations using 74-plex SNPs system [J]. Hereditas(Beijing), 2020, 42(3): 296-308. |
| [9] | Hengzhen Cui, Mizhu Sun, Runzhi Wang, Chenyu Li, Yuxuan Huang, Qiuju Huang, Xiaomeng Qiao. DNA methylation in the medial prefrontal cortex regulates alcohol-related behavior in rats [J]. Hereditas(Beijing), 2020, 42(1): 112-125. |
| [10] | Xinyuan Wang, Yu Zhang, Nan Yang, He Cheng, Yujie Sun. DNMT3a mediates paclitaxel-induced abnormal expression of LINE-1 by increasing the intragenic methylation [J]. Hereditas(Beijing), 2020, 42(1): 100-111. |
| [11] | Xin Huang,Yongqiang Chen,Guoliang Xu,Shuhong Peng. DNA methylation in adipose tissue and the development of diabetes and obesity [J]. Hereditas(Beijing), 2019, 41(2): 98-110. |
| [12] | Xuqiong Yang, Zhenfang Wu, Zicong Li. Advances in epigenetic reprogramming of somatic cells nuclear transfer in mammals [J]. Hereditas(Beijing), 2019, 41(12): 1099-1109. |
| [13] | Yunfeng Pan, Yanyi Wang, Jingwen Chen, Yimei Fan. Mitochondrial metabolism’s effect on epigenetic change and aging [J]. Hereditas(Beijing), 2019, 41(10): 893-904. |
| [14] | Junyi Ju,Quan Zhao. Regulation of γ-globin gene expression and its clinical applications [J]. Hereditas(Beijing), 2018, 40(6): 429-444. |
| [15] | Bi Wu, Wei Hu, Yongzhong Xing. The history and prospect of rice genetic breeding in China [J]. Hereditas(Beijing), 2018, 40(10): 841-857. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||