遗传 ›› 2021, Vol. 43 ›› Issue (8): 758-774.doi: 10.16288/j.yczz.21-131
收稿日期:2021-04-11
修回日期:2021-05-12
出版日期:2021-08-20
发布日期:2021-06-30
作者简介:何象龙,在读硕士研究生,专业方向:生物学。E-mail: 基金资助:
Xianglong He( ), Jinhuan Li, Qiang Wu
), Jinhuan Li, Qiang Wu
Received:2021-04-11
Revised:2021-05-12
Online:2021-08-20
Published:2021-06-30
Supported by:摘要:
CTCF (CCCTC-binding factor)是一种重要的染色质架构蛋白,其与绝缘子的方向性结合在哺乳动物基因组三维空间结构形成和维持中起着至关重要的作用。正向-反向相对方向的CTCF结合位点(简称CTCF位点)可以在染色质黏连蛋白(cohesin)的协助下,形成染色质环,介导远距离DNA元件之间的相互作用;而在染色质拓扑结构域边界区域的CTCF位点呈现反向-正向相背方向分布,发挥绝缘子的功能。为进一步研究CTCF介导染色质环的形成与其绝缘功能之间的关系,本研究采用DNA片段编辑方法通过设计成对sgRNA (dual sgRNA)构建了一系列HOXD基因簇区域CTCF位点反转的单细胞克隆。定量高分辨率染色质构象捕获实验显示边界区域CTCF位点反转会改变原有的染色质环方向,通过环挤出模型(loop extrusion)形成新的染色质环,引起染色质拓扑结构域边界漂移至新形成的一对反向-正向CTCF位点处。此外,串联排列的CTCF位点可以通过阻碍反方向渗透的黏连蛋白继续滑动发挥绝缘子的功能。RNA-seq实验发现CTCF位点反转引起的局部基因组空间结构变化会进一步影响基因的表达。上述研究表明相邻两个染色质拓扑结构域边界区域的反向-正向CTCF位点可以通过与各自所在拓扑结构域内相向的CTCF位点形成染色质环,阻碍黏连蛋白滑动,该发现为进一步研究CTCF的绝缘功能和其对基因组拓扑结构的影响提供了参考。
何象龙, 李金环, 吴强. HOXD基因簇内一系列CTCF位点反转揭示绝缘子功能[J]. 遗传, 2021, 43(8): 758-774.
Xianglong He, Jinhuan Li, Qiang Wu. Combinatorial CRISPR inversions of CTCF sites in HOXD cluster reveal complex insulator function[J]. Hereditas(Beijing), 2021, 43(8): 758-774.
表1
引物序列"
| 类型 | 引物名称 | 序列(5′→3′) |
|---|---|---|
| sgRNA | a-sgRNA1F | accgCGAAGAGTGCGGGAGAACGG |
| a-sgRNA1R | aaacCCGTTCTCCCGCACTCTTCG | |
| b-sgRNA1F | accgGGCGCATCAGGAATGTAAG | |
| b-sgRNA1R | aaacCTTACATTCCTGATGCGCC | |
| b-sgRNA2F | accgTTCCAGAGATTATGAGCCAG | |
| b-sgRNA2R | aaacCTGGCTCATAATCTCTGGAA | |
| c-sgRNA2F | accgAGTAAAACTGGGTGTTGCC | |
| c-sgRNA2R | aaacGGCAACACCCAGTTTTACT | |
| e-sgRNA1F | accgAACTGTGCTCAAACGCTCTC | |
| e-sgRNA1R | aaacGAGAGCGTTTGAGCACAGTT | |
| e-sgRNA2F | accgGAGGCGCAAACAGCTGTTGT | |
| e-sgRNA2R | aaacACAACAGCTGTTTGCGCCTC | |
| PCR | a-1F | TTCCAGCACCTCGGCTTTGTC |
| a-1R | CCCACTTTCCACCTCTGTCCTG | |
| b-1F | GTCCGCCCGTGAGCTTCTGAA | |
| b-1R | GCGTTGGCAGGGCTGGACTC | |
| b-1F1 | TTCCCTGACCCTCCAAGCAC | |
| b-1R1 | CTCACAGCAGCCGAAACCG | |
| b-2F | GACACCTTGGTTCAGGCTCG | |
| b-2R | CAGAGGCTGCATCAAACCAC | |
| c-1F | TGATGCAGCCTCTGTGACCG | |
| c-1R | AGTTTTCCCGTGGCGTCTGA | |
| c-2F | GGACAACCCCTCACCTTGAA | |
| c-2R | TATGGTGCCCAAAGTCCCAC | |
| e-1F | TTCCCTGTCCCAGCTTGATTTC | |
| e-1R | TCAACAGTGAAGGGCGGTGC | |
| e-2F | CAAGCCACTCTCCCGCCACTA | |
| e-2R | TCGCTCTCGTCCTCTCTTGGG | |
| bio-primer | upstream-a-bioprimer | 5′biotin-GGACGAGGGACATAGAGAGTTC |
| downstream-e-bioprimer | 5′biotin- AGGAGCCCAGGCATAGAGAC | |
| CS38-bioprimer | 5′biotin-CCGAATTAAATCCCCGTGAC | |
| Island5-bioprimer | 5′biotin-AAACACAAATGCATCAACCTG | |
| P5 primer | upstream-a-P5 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTCTGTCTAGAGATTCAAGTATTTCCCA |
| P5 primer | downstream-e-P5 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTACGCGATACTCAAGGGAATACAAGCC |
| CS38-P5 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTAGGTTACAGTGTCATTTTCCCTG | |
| Island5-P5 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTATGCATCTCATGAAGCTGGCATCT | |
| P7 index | P7-index-1 | CAAGCAGAAGACGGCATACGAGATCGAGTAATGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT |
| P7-index-2 | CAAGCAGAAGACGGCATACGAGATTCTCCGGAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-3 | CAAGCAGAAGACGGCATACGAGATAATGAGCGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-4 | CAAGCAGAAGACGGCATACGAGATGGAATCTCGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-5 | CAAGCAGAAGACGGCATACGAGATTTCTGAATGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-6 | CAAGCAGAAGACGGCATACGAGATACGAATTCGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-7 | CAAGCAGAAGACGGCATACGAGATAGCTTCAGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-8 | CAAGCAGAAGACGGCATACGAGATGCGCATTAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-9 | CAAGCAGAAGACGGCATACGAGATCATAGCCGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-10 | CAAGCAGAAGACGGCATACGAGATTTCGCGGAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-11 | CAAGCAGAAGACGGCATACGAGATGCGCGAGAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-12 | CAAGCAGAAGACGGCATACGAGATCTATCGCTGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-13 | CAAGCAGAAGACGGCATACGAGATAGAGTACTGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-14 | CAAGCAGAAGACGGCATACGAGATGCTCCGTAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-15 | CAAGCAGAAGACGGCATACGAGATCATGAGAGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-16 | CAAGCAGAAGACGGCATACGAGATTGAATCGCGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-17 | CAAGCAGAAGACGGCATACGAGATGTCTGACTGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-18 | CAAGCAGAAGACGGCATACGAGATCTGAATGCGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-19 | CAAGCAGAAGACGGCATACGAGATCGCTTCTGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-20 | CAAGCAGAAGACGGCATACGAGATTCGCATGAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-21 | CAAGCAGAAGACGGCATACGAGATAATAGCAGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-22 | CAAGCAGAAGACGGCATACGAGATGTCGCGTAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-23 | CAAGCAGAAGACGGCATACGAGATACGCGATAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7 index | P7-index-24 | CAAGCAGAAGACGGCATACGAGATTGATCGATGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT |
| P7-index-25 | CAAGCAGAAGACGGCATACGAGATCCGCATGAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-26 | CAAGCAGAAGACGGCATACGAGATCCACAATCGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-27 | CAAGCAGAAGACGGCATACGAGATGATGTTCGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-28 | CAAGCAGAAGACGGCATACGAGATGAATACGTGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-29 | CAAGCAGAAGACGGCATACGAGATTAGATACCGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-30 | CAAGCAGAAGACGGCATACGAGATTATGGTCGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| adapter | adapter-upper | GACGTGTGCTCTTCCGATCTGNNNNNN-3`NH2 C6 |
| adapter-lower | 5′P-CAGATCGGAAGAGCACACGTC-3`NH2 C6 |

图1
人类HOXD基因簇区域CTCF位点序列分析 A:HOXD基因簇区域基因组三维空间结构示意图。HOXD基因簇位于C-DOM和T-DOM两个TAD边界处,位于HOXD基因簇附近的CBS a-e与C-DOM中的正向CBS形成染色质环,CBS f-g与T-DOM中的成串反向CBS形成染色质环。B:HOXD基因簇区域CBS序列比对图。正向CBS的方向为module 1~4,其中module 1与module 2之间相隔5 bp或者6 bp。DNA序列中的“-”表示该CBS的module 1与module 2间隔为5 bp,故用“-”补齐。右侧CBS方向列,“+”表示CBS为正向,“-”表示该CBS为反向且显示DNA序列是反向互补序列。C:利用CRISPR/Cas9系统进行DNA片段反转示意图。针对预反转片段设计一对sgRNA进行片段编辑,会产生上游接口和下游接口;在切割点附近设计引物,用引物F1+F2、R1+R2进行PCR鉴定,若出现目的条带且F1+R1、F2+R2和F1+R2无目的条带,则为纯合反转单细胞克隆。D:HOXD基因簇内CBS组合性反转示意图,共构建五种反转单细胞克隆株。"
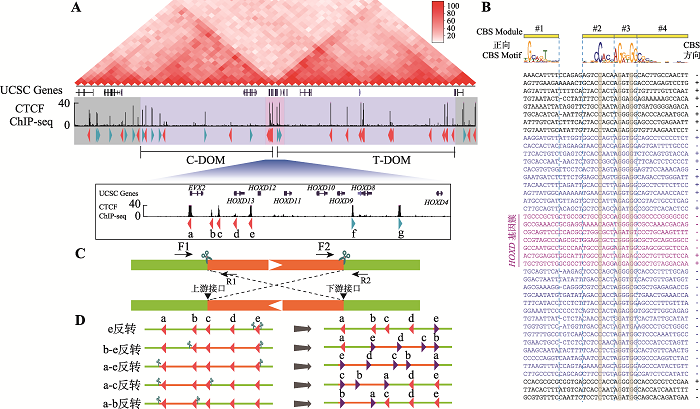
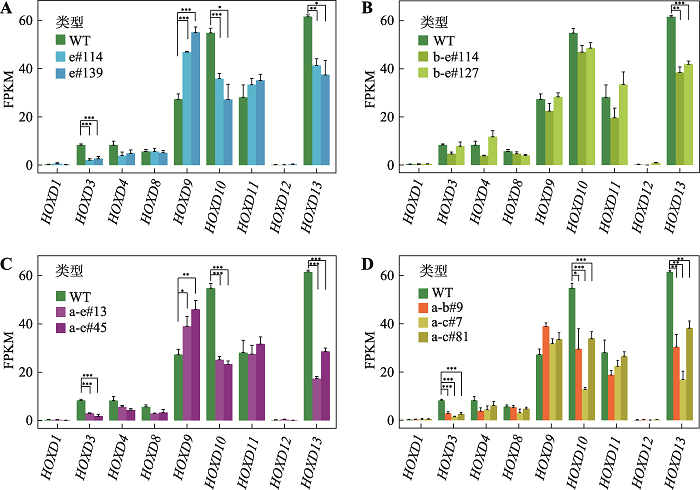
图6
CTCF位点反转改变HOXD基因表达 A:CBS e反转后,HOXD3、HOXD10和HOXD13的表达水平明显降低,HOXD9的表达水平升高;*:P<0.05,**:P<0.01,***:P < 0.001;FPKM,fragments per kilobase of transcript per million mapped reads。B:CBS b-e反转后,HOXD13表达水平显著降低。C:CBS a-e反转后,HOXD3、HOXD10和HOXD13的表达水平降低,而HOXD9的表达水平上升。D:CBS a-b或CBS a-c反转后,HOXD3、HOXD10和HOXD13的表达水平降低。"
| [1] |
Kumar Y, Sengupta D, Bickmore W. Recent advances in the spatial organization of the mammalian genome. J Biosci, 2020, 45:18.
doi: 10.1007/s12038-019-9968-1 |
| [2] |
Wu Q, Liu PF, Wang LY. Many facades of CTCF unified by its coding for three-dimensional genome architecture. J Genet Genomics, 2020, 47(8):407-424.
doi: 10.1016/j.jgg.2020.06.008 |
| [3] | Ning CY, He MN, Tang QZ, Zhu Q, Li MZ, Li DY. Advances in mammalian three-dimensional genome by using Hi-C technology approach. Hereditas(Beijing), 2019, 41(3):215-233. |
| 宁椿游, 何梦楠, 唐茜子, 朱庆, 李明洲, 李地艳. 基于Hi-C技术哺乳动物三维基因组研究进展. 遗传, 2019, 41(3):215-233. | |
| [4] |
Marchal C, Sima J, Gilbert DM. Control of DNA replication timing in the 3D genome. Nat Rev Mol Cell Biol, 2019, 20(12):721-737.
doi: 10.1038/s41580-019-0162-y |
| [5] |
Guo Y, Xu Q, Canzio D, Shou J, Li JH, Gorkin DU, Jung I, Wu HY, Zhai YN, Tang YX, Lu YC, Wu YH, Jia ZL, Li W, Zhang MQ, Ren B, Krainer AR, Maniatis T, Wu Q. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell, 2015, 162(4):900-910.
doi: 10.1016/j.cell.2015.07.038 pmid: 26276636 |
| [6] |
de Laat W, Duboule D. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature, 2013, 502(7472):499-506.
doi: 10.1038/nature12753 |
| [7] |
Sanders JT, Freeman TF, Xu Y, Golloshi R, Stallard MA, Hill AM, San Martin R, Balajee AS, McCord RP. Radiation-induced DNA damage and repair effects on 3D genome organization. Nat Commun, 2020, 11(1):6178.
doi: 10.1038/s41467-020-20047-w pmid: 33268790 |
| [8] |
Arnould C, Rocher V, Finoux AL, Clouaire T, Li K, Zhou F, Caron P, Mangeot PE, Ricci EP, Mourad R, Haber JE, Noordermeer D, Legube G. Loop extrusion as a mechanism for formation of DNA damage repair foci. Nature, 2021, 590(7847):660-665.
doi: 10.1038/s41586-021-03193-z |
| [9] | Cremer T, Cremer M. Chromosome territories. Cold Spring Harb Perspect Biol, 2010, 2(3):a003889. |
| [10] |
Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science, 2002, 295(5558):1306-1311.
pmid: 11847345 |
| [11] |
Zhao ZH, Tavoosidana G, Sjölinder M, Göndör A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, Pant V, Tiwari V, Kurukuti S, Ohlsson R. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet, 2006, 38(11):1341-1347.
doi: 10.1038/ng1891 |
| [12] |
Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science, 2009, 326(5950):289-293.
doi: 10.1126/science.1181369 pmid: 19815776 |
| [13] |
Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, Chew EGY, Huang PYH, Welboren WJ, Han YY, Ooi HS, Ariyaratne PN, Vega VB, Luo YQ, Tan PY, Choy PY, Wansa KDSA, Zhao B, Lim KS, Leow SC, Yow JS, Joseph R, Li HX, Desai KV, Thomsen JS, Lee YK, Karuturi RKM, Herve T, Bourque G, Stunnenberg HG, Ruan XA, Cacheux- Rataboul V, Sung WK, Liu ET, Wei CL, Cheung E, Ruan YJ. An oestrogen-receptor-α-bound human chromatin interactome. Nature, 2009, 462(7269):58-64.
doi: 10.1038/nature08497 |
| [14] |
Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, Gribnau J, Barillot E, Bluthgen N, Dekker J, Heard E. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature, 2012, 485(7398):381-385.
doi: 10.1038/nature11049 |
| [15] |
Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature, 2012, 485(7398):376-380.
doi: 10.1038/nature11082 |
| [16] |
Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, Aiden EL. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell, 2014, 159(7):1665-1680.
doi: 10.1016/j.cell.2014.11.021 |
| [17] |
Guo Y, Monahan K, Wu HY, Gertz J, Varley KE, Li W, Myers RM, Maniatis T, Wu Q. CTCF/cohesin-mediated DNA looping is required for protocadherin α promoter choice. Proc Natl Acad Sci USA, 2012, 109(51):21081-21086.
doi: 10.1073/pnas.1219280110 |
| [18] |
Sanborn AL, Rao SSP, Huang SC, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J, Geeting KP, Gnirke A, Melnikov A, McKenna D, Stamenova EK, Lander ES, Aiden EL. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci USA, 2015, 112(47):E6456-E6465.
doi: 10.1073/pnas.1518552112 |
| [19] |
Andrey G, Montavon T, Mascrez B, Gonzalez F, Noordermeer D, Leleu M, Trono D, Spitz F, Duboule D. A switch between topological domains underliesHoxD genes collinearity in mouse limbs. Science, 2013, 340(6137):1234167.
doi: 10.1126/science.1234167 |
| [20] |
Dixon JR, Gorkin DU, Ren B. Chromatin domains: The unit of chromosome organization. Mol Cell, 2016, 62(5):668-680.
doi: 10.1016/j.molcel.2016.05.018 |
| [21] |
Weintraub AS, Li CH, Zamudio AV, Sigova AA, Hannett NM, Day DS, Abraham BJ, Cohen MA, Nabet B, Buckley DL, Guo YE, Hnisz D, Jaenisch R, Bradner JE, Gray NS, Young RA. YY1 is a structural regulator of enhancer- promoter loops. Cell, 2017, 171(7): 1573-1588.e28.
doi: S0092-8674(17)31317-X pmid: 29224777 |
| [22] | Guo Y, Wu Q. Inversion of CTCF binding sites by DNA fragment editing alters genome topology and enhancer/ promoter functions. Hereditas(Beijing), 2015, 37(10):1073-1074. |
| 郭亚, 吴强. 采用DNA片段编辑技术反转CTCF结合位点改变基因组拓扑结构和增强子与启动子功能. 遗传, 2015, 37(10):1073-1074. | |
| [23] |
Lobanenkov VV, Nicolas RH, Adler VV, Paterson H, Klenova EM, Polotskaja AV, Goodwin GH. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC- motif in the 5'-flanking sequence of the chicken c-myc gene. Oncogene, 1990, 5(12):1743-1753.
pmid: 2284094 |
| [24] |
Yin ML, Wang JY, Wang M, Li XM, Zhang M, Wu Q, Wang YL. Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites. Cell Res, 2017, 27(11):1365-1377.
doi: 10.1038/cr.2017.131 |
| [25] |
Nanni L, Ceri S, Logie C. Spatial patterns of CTCF sites define the anatomy of TADs and their boundaries. Genome Biol, 2020, 21(1):197.
doi: 10.1186/s13059-020-02108-x pmid: 32782014 |
| [26] |
Ghirlando R, Felsenfeld G. CTCF: Making the right connections. Genes Dev, 2016, 30(8):881-891.
doi: 10.1101/gad.277863.116 |
| [27] |
Phillips JE, Corces VG. CTCF: Master weaver of the genome. Cell, 2009, 137(7):1194-1211.
doi: 10.1016/j.cell.2009.06.001 |
| [28] |
de Wit E, Vos ESM, Holwerda SJB, Valdes-Quezada C, Verstegen MJAM, Teunissen H, Splinter E, Wijchers PJ, Krijger PHL, de Laat W. CTCF binding polarity determines chromatin looping. Mol Cell, 2015, 60(4):676-684.
doi: 10.1016/j.molcel.2015.09.023 |
| [29] |
Lu YJ, Shou J, Jia ZL, Wu YH, Li JH, Guo Y, Wu Q. Genetic evidence for asymmetric blocking of higher-order chromatin structure by CTCF/cohesin. Protein Cell, 2019, 10(12):914-920.
doi: 10.1007/s13238-019-00656-y |
| [30] |
Wu Q, Jia ZL. Wiring the brain by clustered protocadherin neural codes. Neurosci Bull, 2021, 37(1):117-131.
doi: 10.1007/s12264-020-00578-4 |
| [31] |
Narendra V, Rocha PP, An DS, Raviram R, Skok JA, Mazzoni EO, Reinberg D. CTCF establishes discrete functional chromatin domains at theHox clusters during differentiation. Science, 2015, 347(6225):1017-1021.
doi: 10.1126/science.1262088 pmid: 25722416 |
| [32] |
Ren G, Jin WF, Cui KR, Rodrigez J, Hu GQ, Zhang ZY, Larson DR, Zhao KJ. CTCF-mediated enhancer-promoter interaction is a critical regulator of cell-to-cell variation of gene expression. Mol Cell, 2017, 67(6): 1049-1058.e6.
doi: 10.1016/j.molcel.2017.08.026 |
| [33] |
Gómez-Marín C, Tena JJ, Acemel RD, López-Mayorga M, Naranjo S, de la Calle-Mustienes E, Maeso I, Beccari L, Aneas I, Vielmas E, Bovolenta P, Nobrega MA, Carvajal J, Gómez-Skarmeta JL. Evolutionary comparison reveals that diverging CTCF sites are signatures of ancestral topological associating domains borders. Proc Natl Acad Sci USA, 2015, 112(24):7542-7547.
doi: 10.1073/pnas.1505463112 |
| [34] |
Jia ZL, Li JW, Ge X, Wu YH, Guo Y, Wu Q. Tandem CTCF sites function as insulators to balance spatial chromatin contacts and topological enhancer-promoter selection. Genome Biol, 2020, 21(1):75.
doi: 10.1186/s13059-020-01984-7 |
| [35] |
Zhang D, Huang P, Sharma M, Keller CA, Giardine B, Zhang HY, Gilgenast TG, Phillips-Cremins JE, Hardison RC, Blobel GA. Alteration of genome folding via contact domain boundary insertion. Nat Genet, 2020, 52(10):1076-1087.
doi: 10.1038/s41588-020-0680-8 |
| [36] | Zhai YN, Xu Q, Guo Y, Wu Q. Characterization of a cluster of CTCF-binding sites in a protocadherin regulatory region. Hereditas(Beijing), 2016, 38(4):323-336. |
| 翟亚男, 许泉, 郭亚, 吴强. 原钙粘蛋白基因簇调控区域中成簇的CTCF结合位点分析. 遗传, 2016, 38(4):323-336. | |
| [37] |
Zhang XF, Zhang Y, Ba ZQ, Kyritsis N, Casellas R, Alt FW. Fundamental roles of chromatin loop extrusion in antibody class switching. Nature, 2019, 575(7782):385-389.
doi: 10.1038/s41586-019-1723-0 |
| [38] |
Xu DF, Ma RS, Zhang JH, Liu ZJ, Wu B, Peng JH, Zhai YN, Gong QG, Shi YY, Wu JH, Wu Q, Zhang ZY, Ruan K. Dynamic nature of CTCF tandem 11 zinc fingers in multivalent recognition of DNA as revealed by NMR spectroscopy. J Phys Chem Lett, 2018, 9(14):4020-4028.
doi: 10.1021/acs.jpclett.8b01440 |
| [39] |
Amândio AR, Lopez-Delisle L, Bolt CC, Mascrez B, Duboule D. A complex regulatory landscape involved in the development of mammalian external genitals. eLife, 2020, 9:e52962.
doi: 10.7554/eLife.52962 |
| [40] |
Bianco S, Annunziatella C, Andrey G, Chiariello AM, Esposito A, Fiorillo L, Prisco A, Conte M, Campanile R, Nicodemi M. Modeling single-molecule conformations of the HoxD region in mouse embryonic stem and cortical neuronal cells. Cell Rep, 2019, 28(6): 1574-1583.e4.
doi: 10.1016/j.celrep.2019.07.013 |
| [41] |
Fabre PJ, Leleu M, Mormann BH, Lopez-Delisle L, Noordermeer D, Beccari L, Duboule D. Large scale genomic reorganization of topological domains at theHoxD locus. Genome Biol, 2017, 18(1):149.
doi: 10.1186/s13059-017-1278-z |
| [42] |
Noordermeer D, Leleu M, Splinter E, Rougemont J, De Laat W, Duboule D. The dynamic architecture of Hox gene clusters. Science, 2011, 334(6053):222-225.
doi: 10.1126/science.1207194 pmid: 21998387 |
| [43] |
Montavon T, Soshnikova N, Mascrez B, Joye E, Thevenet L, Splinter E, de Laat W, Spitz F, Duboule D. A regulatory archipelago controls Hox genes transcription in digits. Cell, 2011, 147(5):1132-1145.
doi: 10.1016/j.cell.2011.10.023 pmid: 22118467 |
| [44] |
Rodríguez-Carballo E, Lopez-Delisle L, Willemin A, Beccari L, Gitto S, Mascrez B, Duboule D. Chromatin topology and the timing of enhancer function at theHoxD locus. Proc Natl Acad Sci USA, 2020, 117(49):31231-31241.
doi: 10.1073/pnas.2015083117 |
| [45] |
Li JH, Shou J, Guo Y, Tang YX, Wu YH, Jia ZL, Zhai YN, Chen ZF, Xu Q, Wu Q. Efficient inversions and duplications of mammalian regulatory DNA elements and gene clusters by CRISPR/Cas9. J Mol Cell Biol, 2015, 7(4):284-298.
doi: 10.1093/jmcb/mjv016 |
| [46] | Li JH, Shou J, Wu Q. DNA fragment editing of genomes by CRISPR/Cas9. Hereditas(Beijing), 2015, 37(10):992-1002. |
| 李金环, 寿佳, 吴强. CRISPR/Cas9系统在基因组DNA片段编辑中的应用. 遗传, 2015, 37(10):992-1002. | |
| [47] | Liu PF, Wu Q. Probing 3D genome by CRISPR/Cas9. Hereditas(Beijing), 2020, 42(1):18-31. |
| 刘沛峰, 吴强. CRISPR/Cas9基因编辑在三维基因组研究中的应用. 遗传, 2020, 42(1):18-31. | |
| [48] |
Shou J, Li JH, Liu YB, Wu Q. Precise and predictable CRISPR chromosomal rearrangements reveal principles of Cas9-mediated nucleotide insertion. Mol Cell, 2018, 71(4): 498-509.e4.
doi: S1097-2765(18)30466-0 pmid: 30033371 |
| [49] | Wang LY, Huang HY, Wu Q. The diversity of DNA fragment editing by CRISPR/Cas9 in highly homologous or repetitive sequences. Hereditas(Beijing), 2017, 39(4):313-325. |
| 汪乐洋, 黄海燕, 吴强. 利用CRISPR/Cas9对基因组中高度同源DNA片段编辑多样性的遗传学研究. 遗传, 2017, 39(4):313-325. | |
| [50] |
Cong L, Ran FA, Cox D, Lin SL, Barretto R, Habib N, Hsu PD, Wu XB, Jiang WY, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas9 systems. Science, 2013, 339(6121):819-823.
doi: 10.1126/science.1231143 pmid: 23287718 |
| [51] |
Wu Q, Shou J. Toward precise CRISPR DNA fragment editing and predictable 3D genome engineering. J Mol Cell Biol, 2021, 12(11):828-856.
doi: 10.1093/jmcb/mjaa060 |
| [52] |
Wang N, Jia ZL, Wu Q. RFX5 regulates gene expression of thePcdhα cluster. Hereditas(Beijing), 2020, 42(8):760-774.
doi: 10.16288/j.yczz.20-184 pmid: 32952112 |
|
王娜, 甲芝莲, 吴强. RFX5调控原钙粘蛋白α基因簇的表达. 遗传, 2020, 42(8):760-774.
doi: 10.16288/j.yczz.20-184 pmid: 32952112 |
|
| [53] |
Wu YH, Jia ZL, Ge X, Wu Q. Three-dimensional genome architectural CCCTC-binding factor makes choice in duplicated enhancers atPcdhα locus. Sci China Life Sci, 2020, 63(6):835-844.
doi: 10.1007/s11427-019-1598-4 |
| [54] |
Zuin J, Dixon JR, van der Reijden MIJA, Ye Z, Kolovos P, Brouwer RWW, van de Corput MPC, van de Werken HJG, Knoch TA, van IJcken WFJ, Grosveld FG, Ren B, Wendt KS. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci USA, 2014, 111(3):996-1001.
doi: 10.1073/pnas.1317788111 |
| [55] |
Servant N, Varoquaux N, Lajoie BR, Viara E, Chen CJ, Vert JP, Heard E, Dekker J, Barillot E. HiC-Pro: An optimized and flexible pipeline for Hi-C data processing. Genome Biol, 2015, 16:259.
doi: 10.1186/s13059-015-0831-x |
| [56] |
Kruse K, Hug CB, Hernández-Rodríguez B, Vaquerizas JM. TADtool: Visual parameter identification for TAD- calling algorithms. Bioinformatics, 2016, 32(20):3190-3192.
pmid: 27318199 |
| [57] |
Bailey TL, Johnson J, Grant CE, Noble WS. The meme suite. Nucleic Acids Res, 2015, 43(W1):W39-W49.
doi: 10.1093/nar/gkv416 |
| [58] | Chen FZ, You LJ, Yang F, Wang LN, Guo XQ, Gao F, Hua C, Tan C, Fang L, Shan RQ, Zeng WJ, Wang B, Wang R, Xu X, Wei XF. CNGBdb: China National GeneBank DataBase. Hereditas(Beijing), 2020, 42(8):799-809. |
| 陈凤珍, 游丽金, 杨帆, 王丽娜, 郭学芹, 高飞, 华聪, 谈聪, 方林, 单日强, 曾文君, 王博, 王韧, 徐讯, 魏晓锋. CNGBdb: 国家基因库生命大数据平台. 遗传, 2020, 42(8):799-809. | |
| [59] | Guo XQ, Chen FZ, Gao F, Li L, Liu K, You LJ, Hua C, Yang F, Liu WL, Peng CH, Wang LN, Yang XX, Zhou FY, Tong JW, Cai J, Li ZY, Wan B, Zhang L, Yang T, Zhang MW, Yang LL, Yang YW, Zeng WJ, Wang B, Wei XF, Xu X. CNSA: a data repository for archiving omics data. Database (Oxford), 2020, 2020: baaa055. |
| [60] |
Shi X, Shou J, Mehryar MM, Li JW, Wang LY, Zhang M, Huang HY, Sun XF, Wu Q. Cas9 has no exonuclease activity resulting in staggered cleavage with overhangs and predictable di- and tri-nucleotide CRISPR insertions without template donor. Cell Discov, 2019, 5:53.
doi: 10.1038/s41421-019-0120-z |
| [61] |
Gligoris TG, Scheinost JC, Burmann F, Petela N, Chan KL, Uluocak P, Beckouët F, Gruber S, Nasmyth K, Löwe J. Closing the cohesin ring: structure and function of its Smc3-kleisin interface. Science, 2014, 346(6212):963-967.
doi: 10.1126/science.1256917 pmid: 25414305 |
| [62] |
Nishiyama T. Cohesion and cohesin-dependent chromatin organization. Curr Opin Cell Biol, 2019, 58:8-14.
doi: S0955-0674(18)30166-2 pmid: 30544080 |
| [1] | 周聪, 周强伟, 成盛, 李国亮. CTCF在介导三维基因组形成及调控基因表达中的研究进展[J]. 遗传, 2021, 43(9): 816-821. |
| [2] | 王玲, 李金环, 黄海燕, 吴强. 串联反向CTCF位点的系列删除揭示增强子调控HOXD基因簇表达的平衡[J]. 遗传, 2021, 43(8): 775-791. |
| [3] | 韦晔, 李科, 卢大儒, 朱化星. CUT&Tag产物回收和建库方法的优化[J]. 遗传, 2021, 43(4): 362-374. |
| [4] | 王娜, 甲芝莲, 吴强. RFX5调控原钙粘蛋白α基因簇的表达[J]. 遗传, 2020, 42(8): 760-774. |
| [5] | 郑晓飞,黄海燕,吴强. 染色质架构蛋白CTCF调控UGT1基因簇的表达[J]. 遗传, 2019, 41(6): 509-523. |
| [6] | 程霄,杨琼,谭镇东,谭娅,蒲红州,赵雪,张顺华,朱砺. 增强子RNA研究现状[J]. 遗传, 2017, 39(9): 784-797. |
| [7] | 谢德健, 师明磊, 张彦, 王天艺, 沈文龙, 叶丙雨, 李平, 何超, 张香媛, 赵志虎. 利用CRISPR/Cas9技术构建CTCF蛋白降解细胞系[J]. 遗传, 2016, 38(7): 651-657. |
| [8] | 翟亚男, 许泉, 郭亚, 吴强. 原钙粘蛋白基因簇调控区域中成簇的CTCF结合位点分析[J]. 遗传, 2016, 38(4): 323-336. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||

www.chinagene.cn
备案号:京ICP备09063187号-4
总访问:,今日访问:,当前在线:
