遗传 ›› 2020, Vol. 42 ›› Issue (1): 18-31.doi: 10.16288/j.yczz.19-246
收稿日期:2019-08-22
修回日期:2019-11-18
出版日期:2020-01-20
发布日期:2019-11-19
通讯作者:
吴强
E-mail:qwu123@gmail.com
作者简介:刘沛峰,博士研究生,专业方向:生物学。E-mail: lpfmail@foxmail.com
基金资助:Received:2019-08-22
Revised:2019-11-18
Online:2020-01-20
Published:2019-11-19
Contact:
Wu Qiang
E-mail:qwu123@gmail.com
Supported by:摘要:
CRISPR/Cas9系统在基因编辑方面具有巨大优势,能够低成本、可编程、方便快捷地用于动物、植物以及微生物的基因组靶向编辑和功能改造。三维基因组学是近年来兴起的一门研究染色质高级结构动态调控及基因组生物学功能的交叉学科。在三维基因组研究中,通常采用对DNA片段进行基因编辑以模拟基因组结构性变异,标记特定DNA片段,进而研究调控元件对于基因调控、细胞分化、组织发生、器官形成、个体发育的影响,最终阐明三维基因组的组装调控机制和生物学功能。因此,CRISPR及其衍生技术为研究三维基因组提供了极好的遗传学工具。本文主要综述了CRISPR片段编辑及其衍生技术在三维基因组调控与功能研究中的应用,以期为后续研究工作提供理论参考以及新的研究思路。
刘沛峰, 吴强. CRISPR/Cas9基因编辑在三维基因组研究中的应用[J]. 遗传, 2020, 42(1): 18-31.
Peifeng Liu, Qiang Wu. Probing 3D genome by CRISPR/Cas9[J]. Hereditas(Beijing), 2020, 42(1): 18-31.
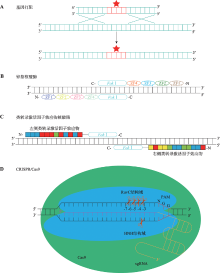
图1
基因组编辑技术 A:基因打靶技术原理。基因打靶技术利用同源重组的方法将目的片段导入基因组。B:锌指核酸酶基因组编辑技术原理。将特异性识别DNA序列的锌指结构串联,与Fok Ⅰ核酸内切酶融合,实现基因组靶向切割。再利用DNA修复系统实现基因组编辑。C:类转录激活因子效应物核酸酶基因组编辑技术原理。将特异性识别DNA序列的类转录激活因子效应物串联,与Fok Ⅰ核酸内切酶融合,实现基因组靶向切割,再利用DNA修复系统实现基因组编辑。D:CRISPR/Cas9基因组编辑技术原理。Cas9蛋白可以被sgRNA引导至基因组特定位点,Cas9不具有核酸外切酶活性,仅仅具有核酸内切酶活性。Cas9的HNH和RuvC结构域分别催化互补链和非互补链的核酸内切反应造成DNA的双链断裂。HNH结构域造成的断裂位点位于PAM序列上游第3与第4个核苷酸之间,而RuvC结构域在PAM序列上游可造成多达4种不同的DNA断裂。"

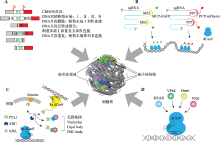
图2
CRISPR在三维基因组研究中的技术方法 A:CRISPR介导的DNA片段编辑。通过一对sgRNA介导Cas9在基因组中进行靶向双位点切割,可以造成基因组片段删除、反转、重复或成环等现象,为三维基因组研究提供模型。B:CRISPR介导的基因组位点可视化。通过在sgRNA上加入MS2与PP7等茎环结构,招募融合了荧光蛋白的MCP、PCP等蛋白,将荧光信号特异性地标记在特定的基因组位点。C:CRISPR介导的基因组位点重定位。利用植物激素ABA介导的PYL1与ABI1的相互作用,将PYL1和ABI1分别结合在不同系统的dCas9或其他核体特有的蛋白上,可以将特定的基因组位点定位到新的目标位点。D:以CRISPR为平台的靶向效应系统。将激活或抑制转录以及表观遗传修饰相关蛋白结合在dCas9上,可以特异性地使基因组特定位点的转录水平以及表观遗传学修饰改变。"
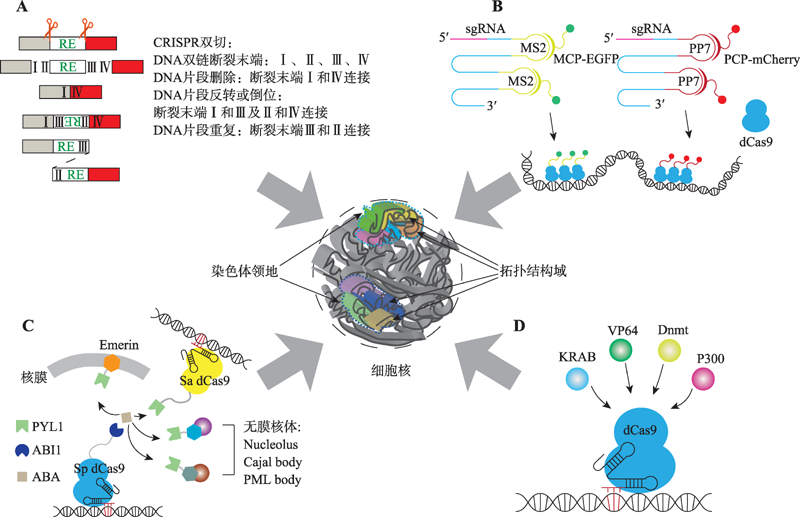
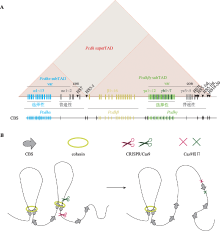
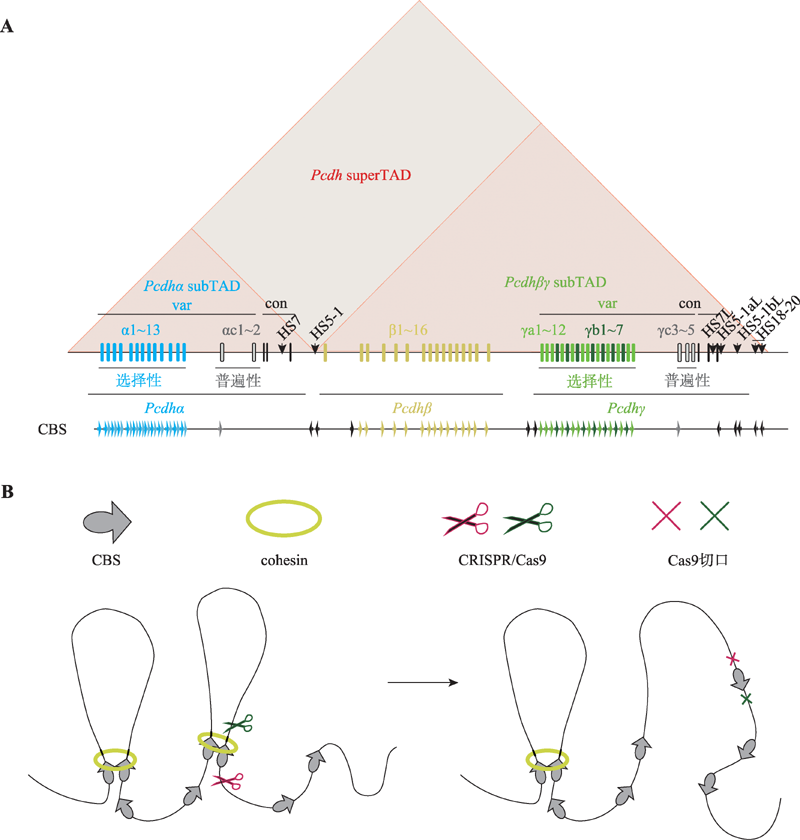
图3
人类原钙粘蛋白基因簇结构以及CRISPR介导的基因组编辑 A:人类原钙粘蛋白基因簇。人类原钙粘蛋白基因座包含α、β、γ 3个基因簇,并且在基因启动子与增强子区域包含大量的CTCF位点(CTCF-binding site, CBS)。通过方向性结合这些CBSs,CTCF可以介导染色质环形成,调控原钙粘蛋白基因表达。B:CRISPR系统介导的基因组元件反转改变染色质拓扑结构。基因组折叠的染色质环挤出模型(chromatin loop extrusion):沿染色质滑动的cohesin可以将染色质挤出形成染色质环,而cohesin的滑动会被一对方向收敛或者说正向-反向(convergent or forward-reverse)的CBS上的CTCF阻滞,这使得染色质环挤出的进程终止,两个CBS在三维空间上被拉近。利用CRISPR/Cas9系统反转CBS方向能够改变染色质环化方向,证明了CTCF位点的方向性在三维基因组折叠中的关键作用。"
| [1] |
Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P . CRISPR provides acquired resistance against viruses in prokaryotes. Science, 2007,315(5819):1709-1712.
doi: 10.1126/science.1138140 pmid: 17379808 |
| [2] |
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E . A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science, 2012,337(6096):816-821.
doi: 10.1126/science.1225829 pmid: 22745249 |
| [3] |
Gasiunas G, Barrangou R, Horvath P, Siksnys V . Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA, 2012,109(39):E2579-E2586.
doi: 10.1073/pnas.1208507109 pmid: 22949671 |
| [4] |
Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F . Genome engineering using the CRISPR-Cas9 system. Nat Protoc, 2013,8(11):2281-2308.
doi: 10.1038/nprot.2013.143 |
| [5] |
Cremer T, Cremer M . Chromosome territories. Cold Spring Harb Perspect Biol, 2010,2(3):a003889.
doi: 10.1101/cshperspect.a003889 pmid: 20300217 |
| [6] |
Tan LZ, Xing D, Chang CH, Li H, Xie XS . Three- dimensional genome structures of single diploid human cells. Science, 2018,361(6405):924-928.
doi: 10.1126/science.aat5641 pmid: 30166492 |
| [7] |
de Laat W, Duboule D . Topology of mammalian developmental enhancers and their regulatory landscapes. Nature, 2013,502(7472):499-506.
doi: 10.1038/nature12753 |
| [8] |
Gibcus JH, Dekker J . The hierarchy of the 3D genome. Mol Cell, 2013,49(5):773-782.
doi: 10.1016/j.molcel.2013.02.011 pmid: 23473598 |
| [9] |
Dixon JR, Gorkin DU, Ren B . Chromatin domains: The unit of chromosome organization. Mol Cell, 2016,62(5):668-680.
doi: 10.1016/j.molcel.2016.05.018 pmid: 27259200 |
| [10] |
Dekker J, Mirny L . The 3D genome as moderator of chromosomal communication. Cell, 2016,164(6):1110-1121.
doi: 10.1016/j.cell.2016.02.007 pmid: 26967279 |
| [11] |
Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU, Jung I, Wu H, Zhai Y, Tang Y, Lu Y, Wu Y, Jia Z, Li W, Zhang MQ, Ren B, Krainer AR, Maniatis T, Wu Q . CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell, 2015,162(4):900-910.
doi: 10.1016/j.cell.2015.07.038 pmid: 26276636 |
| [12] |
Krijger PH, de Laat W . Regulation of disease-associated gene expression in the 3D genome. Nat Rev Mol Cell Biol, 2016,17(12):771-782.
doi: 10.1038/nrm.2016.138 pmid: 27826147 |
| [13] |
Norton HK, Phillips-Cremins JE . Crossed wires: 3D genome misfolding in human disease. . Cell Biol, 2017,216(11):3441-3452.
doi: 10.1083/jcb.201611001 pmid: 28855250 |
| [14] |
Dekker J, Rippe K, Kleckner N . Capturing chromosome conformation. Science, 2002,295(5558):1306-1311.
doi: 10.1126/science.1067799 pmid: 11847345 |
| [15] |
Zhao Z, Tavoosidana G, Sjölinder M, Göndör A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, Pant V, Tiwari V, Kurukuti S, Ohlsson R . Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet, 2006,38(11):1341-1347.
doi: 10.1038/ng1891 pmid: 17033624 |
| [16] |
Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W,. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat Genet, 2006,38(11):1348-1354.
doi: 10.1038/ng1896 pmid: 17033623 |
| [17] |
Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J . Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science, 2009,326(5950):289-293.
doi: 10.1126/science.1181369 pmid: 19815776 |
| [18] |
Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, Chew EG, Huang PY, Welboren WJ, Han Y, Ooi HS, Ariyaratne PN, Vega VB, Luo Y, Tan PY, Choy PY, Wansa KD, Zhao B, Lim KS, Leow SC, Yow JS, Joseph R, Li H, Desai KV, Thomsen JS, Lee YK, Karuturi RK, Herve T, Bourque G, Stunnenberg HG, Ruan X, Cacheux-Rataboul V, Sung WK, Liu ET, Wei CL, Cheung E, Ruan Y . An oestrogen- receptor-alpha-bound human chromatin interactome. Nature, 2009,462(7269):58-64.
doi: 10.1038/nature08497 pmid: 19890323 |
| [19] |
Evans MJ, Kaufman MH . Establishment in culture of pluripotential cells from mouse embryos. Nature, 1981,292(5819):154-156.
doi: 10.1038/292154a0 pmid: 7242681 |
| [20] |
Smithies O, Gregg RG, Boggs SS, Koralewski MA, Kucherlapati RS . Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature, 1985,317(6034):230-234.
doi: 10.1038/317230a0 pmid: 2995814 |
| [21] |
Thomas KR, Folger KR, Capecchi MR . High frequency targeting of genes to specific sites in the mammalian genome. Cell, 1986,44(3):419-428.
doi: 10.1016/0092-8674(86)90463-0 pmid: 3002636 |
| [22] |
Kim YG, Cha J, Chandrasegaran S . Hybrid restriction enzymes: Zinc finger fusions to FokⅠ cleavage domain. Proc Natl Acad Sci USA, 1996,93(3):1156-1160.
doi: 10.1073/pnas.93.3.1156 pmid: 8577732 |
| [23] |
Bibikova M, Carroll D, Segal DJ, Trautman JK, Smith J, Kim YG, Chandrasegaran S . Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol Cell Biol, 2001,21(1):289-297.
doi: 10.1128/MCB.21.1.289-297.2001 pmid: 11113203 |
| [24] |
Moscou MJ, Bogdanove AJ . A simple cipher governs DNA recognition by TAL effectors. Science, 2009,326(5959):1501.
doi: 10.1126/science.1178817 pmid: 19933106 |
| [25] |
Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U . Breaking the code of DNA binding specificity of TAL-type Ⅲ effectors. Science, 2009,326(5959):1509-1512.
doi: 10.1126/science.1178811 pmid: 19933107 |
| [26] |
Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF . Targeting DNA double-strand breaks with TAL effector nucleases. Genetics, 2010,186(2):757-761.
doi: 10.1534/genetics.110.120717 pmid: 20660643 |
| [27] |
Li J, Zhang Y, Chen KL, Shan QW, Wang YP, Liang Z, Gao CX . CRISPR/Cas: A novel way of RNA-guided genome editing. Hereditas(Beijing), 2013,35(11):1265-1273.
doi: 10.3724/SP.J.1005.2013.01265 |
|
李君, 张毅, 陈坤玲, 单奇伟, 王延鹏, 梁振, 高彩霞 . CRISPR/Cas系统: RNA靶向的基因组定向编辑新技术. 遗传, 2013,35(11):1265-1273.
doi: 10.3724/SP.J.1005.2013.01265 |
|
| [28] |
Huang H, Wu Q . CRISPR double cutting through the labyrinthine architecture of 3D genomes. . Genet Genomics, 2016,43(5):273-288.
doi: 10.1016/j.jgg.2016.03.006 pmid: 27210040 |
| [29] |
Li JH, Shou J, Guo Y, Tang YX, Wu YH, Jia ZL, Zhai YA, Chen ZF, Xu Q, Wu Q . Efficient inversions and duplications of mammalian regulatory DNA elements and gene clusters by CRISPR/Cas9. . Mol Cell Biol, 2015,7(4):284-298.
doi: 10.1093/jmcb/mjv016 pmid: 25757625 |
| [30] |
Li JH, Shou J, Wu Q . DNA fragment editing of genomes by CRISPR/Cas9. Hereditas(Beijing), 2015,37(10):992-1002.
doi: 10.16288/j.yczz.15-291 pmid: 26496751 |
|
李金环, 寿佳, 吴强 . CRISPR/Cas9系统在基因组DNA片段编辑中的应用. 遗传, 2015,37(10):992-1002.
doi: 10.16288/j.yczz.15-291 pmid: 26496751 |
|
| [31] | Wang LY, Huang HY, Wu Q . The diversity of DNA fragment editing by CRISPR/Cas9 in highly homologous or repetitive sequences. Hereditas(Beijing), 2017,39(4):313-325. |
| 汪乐洋, 黄海燕, 吴强 . 利用CRISPR/Cas9对基因组中高度同源DNA片段编辑多样性的遗传学研究. 遗传, 2017,39(4):313-325. | |
| [32] |
Long CZ . God does not play dice, and neither does CRISPR/Cas9. Natl Sci Rev, 2019,6(3):393-393.
doi: 10.1007/s10943-020-00982-0 pmid: 31953788 |
| [33] |
Shou J, Li J, Liu Y, Wu Q. Precise and predictable CRISPR chromosomal rearrangements reveal principles of Cas9-mediated nucleotide insertion. Mol Cell, 2018,71(4): 498-509.e4.
doi: 10.1016/j.molcel.2018.06.021 pmid: 30033371 |
| [34] |
McVey M, Lee SE . MMEJ repair of double-strand breaks (director's cut): Deleted sequences and alternative endings. Trends Genet, 2008,24(11):529-538.
doi: 10.1016/j.tig.2008.08.007 pmid: 18809224 |
| [35] |
Allen F, Crepaldi L, Alsinet C, Strong AJ, Kleshchevnikov V, De Angeli P, Páleníková P, Khodak A, Kiselev V, Kosicki M, Bassett AR, Harding H, Galanty Y, Muñoz- Martínez F, Metzakopian E, Jackson SP, Parts L . Predicting the mutations generated by repair of Cas9-induced double- strand breaks. Nat Biotechnol, 2018,37(1):64-72
doi: 10.1038/nbt.4317 pmid: 30480667 |
| [36] |
Shen MW, Arbab M, Hsu JY, Worstell D, Culbertson SJ, Krabbe O, Cassa CA, Liu DR, Gifford DK, Sherwood RI . Predictable and precise template-free CRISPR editing of pathogenic variants. Nature, 2018,563(7733):646-651.
doi: 10.1038/s41586-018-0686-x pmid: 30405244 |
| [37] |
Shi X, Shou J, Mehryar MM, Li JW, Wang LY, Zhang M, Huang HY, Sun XF, Wu Q . Cas9 has no exonuclease activity resulting in staggered cleavage with overhangs and predictable di- and tri-nucleotide CRISPR insertions without template donor. Cell Discov, 2019,5:53.
doi: 10.1038/s41421-019-0120-z pmid: 31636963 |
| [38] |
Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, Zhang LN, Weintraub AS, Schujiers J, Lee TI, Zhao K, Young RA . Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell, 2014,159(2):374-387.
doi: 10.1016/j.cell.2014.09.030 |
| [39] |
Despang A, Schöpflin R, Franke M, Ali S, Jerković I, Paliou C, Chan WL, Timmermann B, Wittler L, Vingron M, Mundlos S, Ibrahim DM . Functional dissection of the Sox9-Kcnj2 locus identifies nonessential and instructive roles of TAD architecture. Nat Genet, 2019,51(8):1263-1271.
doi: 10.1038/s41588-019-0466-z pmid: 31358994 |
| [40] |
Kragesteen BK, Spielmann M, Paliou C, Heinrich V, Schöpflin R, Esposito A, Annunziatella C, Bianco S, Chiariello AM, Jerković I, Harabula I, Guckelberger P, Pechstein M, Wittler L, Chan WL, Franke M, Lupiáñez DG, Kraft K, Timmermann B, Vingron M, Visel A, Nicodemi M, Mundlos S, Andrey G . Dynamic 3D chromatin architecture contributes to enhancer specificity and limb morphogenesis. Nat Genet, 2018,50(10):1463-1473.
doi: 10.1038/s41588-018-0221-x pmid: 30262816 |
| [41] |
Sima J, Chakraborty A, Dileep V, Michalski M, Klein KN, Holcomb NP, Turner JL, Paulsen MT, Rivera-Mulia JC, Trevilla-Garcia C, Bartlett DA, Zhao PA, Washburn BK, Nora EP, Kraft K, Mundlos S, Bruneau BG, Ljungman M, Fraser P, Ay F, Gilbert DM. Identifying cis elements for spatiotemporal control of mammalian DNA replication. Cell, 2019, 176(4): 816-830.e18.
doi: 10.1016/j.cell.2018.11.036 pmid: 30595451 |
| [42] |
Zuo E, Huo X, Yao X, Hu X, Sun Y, Yin J, He B, Wang X, Shi L, Ping J, Wei Y, Ying W, Wei W, Liu W, Tang C, Li Y, Hu J, Yang H . CRISPR/Cas9-mediated targeted chromosome elimination. Genome Biol, 2017,18(1):224.
doi: 10.1186/s13059-017-1354-4 pmid: 29178945 |
| [43] |
Shao Y, Lu N, Wu Z, Cai C, Wang S, Zhang LL, Zhou F, Xiao S, Liu L, Zeng X, Zheng H, Yang C, Zhao Z, Zhao G, Zhou JQ, Xue X, Qin Z . Creating a functional single- chromosome yeast. Nature, 2018,560(7718):331-335.
doi: 10.1038/s41586-018-0382-x pmid: 30069045 |
| [44] |
Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, Park J, Blackburn EH, Weissman JS, Qi LS, Huang B . Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell, 2013,155(7):1479-1491.
doi: 10.1016/j.cell.2013.12.001 |
| [45] |
Deng W, Shi X, Tjian R, Lionnet T, Singer RH . Casfish: CRISPR/Cas9-mediated in situ labeling of genomic loci in fixed cells. Proc Natl Acad Sci USA, 2015,112(38):11870-11875.
doi: 10.1073/pnas.1515692112 pmid: 26324940 |
| [46] |
Ma H, Naseri A, Reyes-Gutierrez P, Wolfe SA, Zhang S, Pederson T . Multicolor CRISPR labeling of chromosomal loci in human cells. Proc Natl Acad Sci USA, 2015,112(10):3002-3007.
doi: 10.1073/pnas.1420024112 pmid: 25713381 |
| [47] |
Chen B, Hu J, Almeida R, Liu H, Balakrishnan S, Covill-Cooke C, Lim WA, Huang B . Expanding the CRISPR imaging toolset with Staphylococcus aureus Cas9 for simultaneous imaging of multiple genomic loci. Nucleic Acids Res, 2016,44(8):e75.
doi: 10.1093/nar/gkv1533 pmid: 26740581 |
| [48] |
Fu Y, Rocha PP, Luo VM, Raviram R, Deng Y, Mazzoni EO, Skok JA . CRISPR-dCas9 and sgRNA scaffolds enable dual-colour live imaging of satellite sequences and repeat- enriched individual loci. Nat Commun, 2016,7:11707.
doi: 10.1038/ncomms11707 pmid: 27222091 |
| [49] |
Maass PG, Barutcu AR, Weiner CL, Rinn JL. Inter- chromosomal contact properties in live-cell imaging and in Hi-C. Mol Cell, 2018, 69(6): 1039-1045.e3.
doi: 10.1016/j.molcel.2018.02.007 pmid: 29526697 |
| [50] |
Ma H, Tu LC, Naseri A, Huisman M, Zhang S, Grunwald D, Pederson T . Multiplexed labeling of genomic loci with dCas9 and engineered sgRNAs using CRISPRainbow. Nat Biotechnol, 2016,34(5):528-530.
doi: 10.1038/nbt.3526 pmid: 27088723 |
| [51] |
Shao S, Zhang W, Hu H, Xue B, Qin J, Sun C, Sun Y, Wei W, Sun Y . Long-term dual-color tracking of genomic loci by modified sgRNAs of the CRISPR/Cas9 system. Nucleic Acids Res, 2016,44(9):e86.
doi: 10.1093/nar/gkw066 pmid: 26850639 |
| [52] |
Qin P, Parlak M, Kuscu C, Bandaria J, Mir M, Szlachta K, Singh R, Darzacq X, Yildiz A, Adli M . Live cell imaging of low- and non-repetitive chromosome loci using CRISPR-Cas9. Nat Commun, 2017,8:14725.
doi: 10.1038/ncomms14725 pmid: 28290446 |
| [53] |
Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, Ylstra B, Chubb JR, Bickmore WA . Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet, 2008,4(3):e1000039.
doi: 10.1371/journal.pgen.1000039 pmid: 18369458 |
| [54] |
Kumaran RI, Spector DL . A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. . Cell Biol, 2008,180(1):51-65.
doi: 10.1083/jcb.200706060 pmid: 18195101 |
| [55] |
Wang H, Xu X, Nguyen CM, Liu Y, Gao Y, Lin X, Daley T, Kipniss NH, La Russa M, Qi LS. CRISPR-mediated programmable 3D genome positioning and nuclear organization. Cell, 2018, 175(5): 1405-1417.e14.
doi: 10.1016/j.cell.2018.09.013 pmid: 30318144 |
| [56] |
Morgan SL, Mariano NC, Bermudez A, Arruda NL, Wu FT, Luo YH, Shankar G, Jia L, Chen HL, Hu JF, Hoffman AR, Huang CC, Pitteri SJ, Wang KC . Manipulation of nuclear architecture through CRISPR-mediated chromosomal looping. Nat Commun, 2017,8:15993.
doi: 10.1038/ncomms15993 pmid: 28703221 |
| [57] |
Cheng AW, Wang HY, Yang H, Shi YL, Katz Y, Theunissen TW, Rangarajan S, Shivalila CS, Dadon DB, Jaenisch R . Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res, 2013,23(10):1163-1171.
doi: 10.1038/cr.2013.122 |
| [58] |
Black JB, Adler AF, Wang HG , D'Ippolito AM, Hutchinson HA, Reddy TE, Pitt GS, Leong KW, Gersbach CA. Targeted epigenetic remodeling of endogenous loci by CRISPR/ Cas9-based transcriptional activators directly converts fibroblasts to neuronal cells. Cell Stem Cell, 2016,19(3):406-414.
doi: 10.1016/j.stem.2016.07.001 pmid: 27524438 |
| [59] |
Himeda CL, Jones TI, Jones PL . CRISPR/dCas9-mediated transcriptional inhibition ameliorates the epigenetic dysregulation at D4Z4 and represses DUX4-fl in FSH muscular dystrophy. Mol Ther, 2016,24(3):527-535.
doi: 10.1038/mt.2015.200 pmid: 26527377 |
| [60] | Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, Shu J, Dadon D, Young RA , Jaenisch R. Editing DNA methylation in the mammalian genome. Cell, 2016, 167(1): 233- 247. e17. |
| [61] |
Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR . Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature, 2016,533(7603):420-424.
doi: 10.1038/nature17946 pmid: 27096365 |
| [62] | Liao HK, Hatanaka F, Araoka T, Reddy P, Wu MZ, Sui Y, Yamauchi T, Sakurai M, O'Keefe DD, Núñez-Delicado E, Guillen P, Campistol JM, Wu CJ, Lu LF, Esteban CR , Izpisua Belmonte JC. In vivo target gene activation via CRISPR/Cas9-mediated trans-epigenetic modulation. Cell, 2017, 171(7): 1495- 1507. e15. |
| [63] |
Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA . Repurposing CRISPR as an RNA- guided platform for sequence-specific control of gene expression. Cell, 2013,152(5):1173-1183.
doi: 10.1016/j.cell.2013.02.022 |
| [64] |
Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS . CRISPR- mediated modular RNA-guided regulation of transcription in eukaryotes. Cell, 2013,154(2):442-451.
doi: 10.1016/j.cell.2013.06.044 pmid: 23849981 |
| [65] |
Gao Y, Xiong X, Wong S, Charles EJ, Lim WA, Qi LS . Complex transcriptional modulation with orthogonal and inducible dCas9 regulators. Nat Methods, 2016,13(12):1043-1049.
doi: 10.1038/nmeth.4042 pmid: 27776111 |
| [66] |
Zuo E, Sun YD, Wei W, Yuan TL, Ying WQ, Sun H, Yuan LY, Steinmetz LM, Li YX, Yang H . Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science, 2019,364(6437):289-292.
doi: 10.1126/science.aav9973 pmid: 30819928 |
| [67] |
Jin S, Zong Y, Gao Q, Zhu ZX, Wang YP, Qin P, Liang CZ, Wang DW, Qiu JL, Zhang F, Gao CX . Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science, 2019,364(6437):292-295.
doi: 10.1126/science.aaw7166 pmid: 30819931 |
| [68] |
Hilton IB , D'Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol, 2015,33(5):510-517.
doi: 10.1038/nbt.3199 pmid: 25849900 |
| [69] |
Kearns NA, Pham H, Tabak B, Genga RM, Silverstein NJ, Garber M, Maehr R . Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods, 2015,12(5):401-403.
doi: 10.1038/nmeth.3325 pmid: 25775043 |
| [70] |
Wang H, Guo R, Du Z, Bai L, Li L, Cui J, Li W, Hoffman AR, Hu JF . Epigenetic targeting of Granulin in hepatoma cells by synthetic CRISPR dCas9 epi-suppressors. Mol Ther Nucleic Acids, 2018,11:23-33.
doi: 10.1016/j.ecoenv.2019.110160 pmid: 31951899 |
| [71] |
Wang G, Chow R D, Bai Z, Zhu L, Errami Y, Dai X, Dong M B, Ye L, Zhang X, Renauer P A, Park J J, Shen L, Ye H, Fuchs C S, Chen S . Multiplexed activation of endogenous genes by CRISPRa elicits potent antitumor immunity. Nat Immunol, 2019,20:1494-1505.
doi: 10.1038/s41590-019-0500-4 pmid: 31611701 |
| [72] |
Crane E, Bian Q , McCord RP, Lajoie BR, Wheeler BS, Ralston EJ, Uzawa S, Dekker J, Meyer BJ. Condensin- driven remodeling of x-chromosome topology during dosage compensation. Nature, 2015,523(7559):240-244.
doi: 10.1038/nature14450 pmid: 26030525 |
| [73] | Gasperini M, Hill AJ, McFaline-Figueroa JL, Martin B, Kim S, Zhang MD, Jackson D, Leith A, Schreiber J, Noble WS, Trapnell C, Ahituv N , Shendure J. A genome-wide framework for mapping gene regulation via cellular genetic screens. Cell, 2019, 176(1-2): 377- 390. e19. |
| [74] |
de Wit E, Vos ES, Holwerda SJ, Valdes-Quezada C, Verstegen MJ, Teunissen H, Splinter E, Wijchers PJ, Krijger PH, de Laat W . CTCF binding polarity determines chromatin looping. Mol Cell, 2015,60(4):676-684.
doi: 10.1016/j.molcel.2015.09.023 pmid: 26527277 |
| [75] |
Sanborn AL, Rao SS, Huang SC, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J, Geeting KP, Gnirke A, Melnikov A , McKenna D, Stamenova EK, Lander ES, Aiden EL. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci USA, 2015,112(47):E6456-6465.
doi: 10.1073/pnas.1518552112 pmid: 26499245 |
| [76] |
Zheng XF, Huang HY, Wu Q . Chromatin architectural protein CTCF regulates gene expression of the UGT1 cluster. Hereditas(Beijing), 2019,41(6):509-523.
doi: 10.16288/j.yczz.19-072 pmid: 31257199 |
|
郑晓飞, 黄海燕, 吴强 . 染色质架构蛋白CTCF调控UGT1基因簇的表达. 遗传, 2019,41(6):509-523.
doi: 10.16288/j.yczz.19-072 pmid: 31257199 |
|
| [77] |
Wu Q, Guo Y, Lu YJ, Li JW, Wu YH , Jia ZL. Tandem directional CTCF sites balance protocadherin promoter usage. BioRxiv, 2019, 10. 1101/525543
pmid: 10 |
| [78] |
Lupiáñez D G, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, Santos-Simarro F, Gilbert-Dussardier B, Wittler L, Borschiwer M, Haas SA, Osterwalder M, Franke M, Timmermann B, Hecht J, Spielmann M, Visel A, Mundlos S . Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell, 2015,161(5):1012-1025.
doi: 10.1016/j.cell.2015.04.004 pmid: 25959774 |
| [79] |
Hnisz D, Weintraub AS, Day DS, Valton AL, Bak RO, Li CH, Goldmann J, Lajoie BR, Fan ZP, Sigova AA, Reddy J, Borges-Rivera D, Lee TI, Jaenisch R, Porteus MH, Dekker J, Young RA . Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science, 2016,351(6280):1454-1458.
doi: 10.1126/science.aad9024 pmid: 26940867 |
| [80] |
Guo Y, Monahan K, Wu H, Gertz J, Varley KE, Li W, Myers RM, Maniatis T, Wu Q . CTCF/cohesin-mediated DNA looping is required for protocadherin α promoter choice. Proc Natl Acad Sci USA, 2012,109(51):21081-21086.
doi: 10.1073/pnas.1219280110 pmid: 23204437 |
| [81] | Zhai YN, Xu Q, Guo Y, Wu Q . Characterization of a cluster of CTCF-binding sites in a protocadherin regulatory region. Hereditas(Beijing), 2016,38(4):323-336. |
| 翟亚男, 许泉, 郭亚, 吴强 . 原钙粘蛋白基因簇调控区域中成簇的CTCF结合位点分析. 遗传, 2016,38(4):323-336. | |
| [82] |
Wu Q, Maniatis T . A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell, 1999,97(6):779-790.
doi: 10.1016/s0092-8674(00)80789-8 pmid: 10380929 |
| [83] |
Guo Y, Wu Q . Inversion of CTCF binding sites by DNA fragment editing alters genome topology and enhancer/ promoter functions. Hereditas(Beijing), 2015,37(10):1073-1074.
doi: 10.13703/j.0255-2930.2017.10.012 pmid: 29354976 |
|
郭亚, 吴强 . 采用DNA片段编辑技术反转CTCF结合位点改变基因组拓扑结构和增强子与启动子功能. 遗传, 2015,37(10):1073-1074.
doi: 10.13703/j.0255-2930.2017.10.012 pmid: 29354976 |
|
| [84] |
Lu YJ, Shou J, Jia ZL, Wu YH, Li JH, Guo Y, Wu Q . Genetic evidence for asymmetric blocking of higher-order chromatin structure by CTCF/cohesin. Protein Cell, 2019, 1-7.
doi: 10.1007/s13238-019-0623-2 pmid: 31037510 |
| [85] |
Kleinstiver BP, Sousa AA, Walton RT, Tak YE, Hsu JY, Clement K, Welch MM, Horng JE, Malagon-Lopez J, Scarfò I, Maus MV, Pinello L, Aryee MJ, Joung JK . Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat Biotechnol, 2019,37(3):276-282.
doi: 10.1038/s41587-018-0011-0 pmid: 30742127 |
| [86] |
Liu L, Li X, Ma J, Li Z, You L, Wang J, Wang M, Zhang X, Wang Y. The molecular architecture for RNA-guided RNA cleavage by Cas13a. Cell, 2017, 170(4): 714-726.e10.
doi: 10.1016/j.cell.2017.06.050 pmid: 28757251 |
| [87] |
Ghavi-Helm Y, Jankowski A, Meiers S, Viales RR, Korbel JO, Furlong EEM . Highly rearranged chromosomes reveal uncoupling between genome topology and gene expression. Nat Genet, 2019,51(8):1272-1282.
doi: 10.1038/s41588-019-0462-3 pmid: 31308546 |
| [1] | 王舜泽, 江丰, 朱东丽, 杨铁林, 郭燕. Hi-C技术在三维基因组学和疾病致病机理研究中的应用[J]. 遗传, 2023, 45(4): 279-294. |
| [2] | 陈秀丽, 黄海燕, 吴强. 靶向敲除β-珠蛋白基因座控制区增强子HS2对K562细胞转录组的影响[J]. 遗传, 2022, 44(9): 783-797. |
| [3] | 周聪, 周强伟, 成盛, 李国亮. CTCF在介导三维基因组形成及调控基因表达中的研究进展[J]. 遗传, 2021, 43(9): 816-821. |
| [4] | 罗鑫, 宿兵. 三维基因组分析点亮人类大脑进化之谜[J]. 遗传, 2021, 43(2): 105-107. |
| [5] | 张雨, 方玉达. Cohesin结构及功能研究进展[J]. 遗传, 2020, 42(1): 57-72. |
| [6] | 杨科, 薛征, 吕湘. 细胞终末分化过程中三维基因组结构与功能调控的分子机制[J]. 遗传, 2020, 42(1): 32-44. |
| [7] | 郑晓飞,黄海燕,吴强. 染色质架构蛋白CTCF调控UGT1基因簇的表达[J]. 遗传, 2019, 41(6): 509-523. |
| [8] | 宁椿游,何梦楠,唐茜子,朱庆,李明洲,李地艳. 基于Hi-C技术哺乳动物三维基因组研究进展[J]. 遗传, 2019, 41(3): 215-233. |
| [9] | 汪乐洋,黄海燕,吴强. 利用CRISPR/Cas9对基因组中高度同源DNA片段编辑多样性的遗传学研究[J]. 遗传, 2017, 39(4): 313-325. |
| [10] | 张道微, 张超凡, 董芳, 黄艳岚, 张亚, 周虹. CRISPR/Cas9系统在培育抗病毒植物新种质中的应用[J]. 遗传, 2016, 38(9): 811-820. |
| [11] | 谢胜松,张懿,张利生,李广磊,赵长志,倪攀,赵书红. CRISPR/Cas9系统中sgRNA设计与脱靶效应评估[J]. 遗传, 2015, 37(11): 1125-1136. |
| [12] | 李金环, 寿佳, 吴强. CRISPR/Cas9系统在基因组DNA片段编辑中的应用[J]. 遗传, 2015, 37(10): 992-291. |
| [13] | 璩良, 李华善, 姜运涵, 董春升. CRISPR/Cas9系统的分子机制及其在人类疾病基因治疗中的应用[J]. 遗传, 2015, 37(10): 974-982. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||

www.chinagene.cn
备案号:京ICP备09063187号-4
总访问:,今日访问:,当前在线:

