遗传 ›› 2023, Vol. 45 ›› Issue (5): 447-458.doi: 10.16288/j.yczz.23-001
• 研究报告 • 上一篇
高智慧1,2,3( ), 黄佳新1,2,3, 罗昊玉1,2,3, 徐海冬1,2,3, 娄明1,2,3, 宁博林1,2,3, 邢晓旭1,2,3, 牟芳1,2,3, 李辉1,2,3, 王宁1,2,3(
), 黄佳新1,2,3, 罗昊玉1,2,3, 徐海冬1,2,3, 娄明1,2,3, 宁博林1,2,3, 邢晓旭1,2,3, 牟芳1,2,3, 李辉1,2,3, 王宁1,2,3( )
)
收稿日期:2023-01-03
修回日期:2023-04-04
出版日期:2023-05-20
发布日期:2023-04-19
作者简介:高智慧,在读硕士研究生,专业方向:动物遗传育种与繁殖。E-mail: 基金资助:
Zhihui Gao1,2,3( ), Jiaxin Huang1,2,3, Haoyu Luo1,2,3, Haidong Xu1,2,3, Ming Lou1,2,3, Bolin Ning1,2,3, Xiaoxu Xing1,2,3, Fang Mu1,2,3, Hui Li1,2,3, Ning Wang1,2,3(
), Jiaxin Huang1,2,3, Haoyu Luo1,2,3, Haidong Xu1,2,3, Ming Lou1,2,3, Bolin Ning1,2,3, Xiaoxu Xing1,2,3, Fang Mu1,2,3, Hui Li1,2,3, Ning Wang1,2,3( )
)
Received:2023-01-03
Revised:2023-04-04
Published:2023-05-20
Online:2023-04-19
Supported by:摘要:
神经调节蛋白4(neuregulin 4,NRG4)是一个重要的脂肪细胞因子,在维持哺乳动物能量平衡、调节糖脂代谢和预防非酒精性脂肪性肝病中起着非常重要的作用。目前,人NRG4基因的基因组结构、转录异构体和蛋白异构体等已有深入的研究。本实验室前期研究显示,鸡脂肪组织也表达NRG4,但是目前有关鸡NRG4(chicken NRG4,cNRG4)基因的基因组结构、转录异构体和蛋白异构体尚不清楚。为此,本研究采用RACE(rapid-amplification of cDNA ends)和RT-PCR(reverse transcription-PCR)等技术,系统开展了cNRG4基因的基因组和转录本的结构分析。结果发现,cNRG4基因编码区很小,但该基因却非常复杂,存在选择性转录起始位点、选择性拼接、内含子滞留、隐匿外显子和选择性多聚腺苷酸化,这导致cNRG4基因产生4种不同的5?UTR异构体(cNRG4 A、cNRG4 B、cNRG4 C和cNRG4 D)和6种不同的3?UTR异构体(cNRG4 a、cNRG4 b、cNRG4 c、cNRG4 d、cNRG4 e和cNRG4 f)。基因组结构分析发现,cNRG4基因跨越基因组21,969 bp (Chr.10: 3,490,314~3,512,282),由11个外显子和10个内含子构成。与NCBI数据库中的cNRG4基因mRNA序列(NM_001030544.4)相比,本研究发现了cNRG4基因的2个新外显子和1个隐匿外显子。生物信息学分析、RT-PCR、克隆和测序分析发现,cNRG4基因能编码3种蛋白异构体(cNRG4-1、cNRG4-2和 cNRG4-3)。本研究为进一步开展cNRG4基因的功能和调控研究奠定了基础。
高智慧, 黄佳新, 罗昊玉, 徐海冬, 娄明, 宁博林, 邢晓旭, 牟芳, 李辉, 王宁. 鸡NRG4基因组及转录本结构分析[J]. 遗传, 2023, 45(5): 447-458.
Zhihui Gao, Jiaxin Huang, Haoyu Luo, Haidong Xu, Ming Lou, Bolin Ning, Xiaoxu Xing, Fang Mu, Hui Li, Ning Wang. Characterization of the genomic and transcriptional structure of chicken NRG4 gene[J]. Hereditas(Beijing), 2023, 45(5): 447-458.
表1
本研究所用引物信息"
| 引物类型 | 引物名称 | 引物序列(5?→3?) |
|---|---|---|
| RACE | GSP1-R1 | AGAAGGAGAAGTGAAGCCAAGAGAACTG |
| GSP2-F1 | GTTTGCAGTTCTCTTGGCTTCACTTCTC | |
| RT-PCR | E76-F/E4-R2 | GCCGAGCCAATGGGCGACG GATCCATAACTGGTGCCACAGAGT |
| E85-F/E4-R1 | GGTGAGGCGCGAGAG GACAAAAAGATCCATAACTGGTGC | |
| E56-F/E4-R2 | CTCCTTCACAGGCACAGTTCAG GATCCATAACTGGTGCCACAGAGT | |
| I1-F/E4-R1 | CTCTGTTAACGTTCCGCCTG GACAAAAAGATCCATAACTGGTGC | |
| E4-F /I5-R | TTCCTACTGTACCCAGTCCATTC CTACTTGATTAAATAGCTTCAGCAC | |
| E5-F /I7-R | CTTCTCGGAGTTCTTTTAATTGGA TCTCTAAGATAAAAGTAGGCCTC | |
| CDS1-F/R | ATGCGAACAGATCATGAAG TTACCTGCAAAGGAAGTAG | |
| CDS2-F/R | ATGCGAACAGATCATGAAG TTACTCCGTTATTATTTTGTTGCAG | |
| CDS3-F/R | ATGCGAACAGATCATGAAG TAGTACATGATTACCAGAAGCAA |

图3
鸡NRG4基因新外显子和内含子滞留的RT-PCR鉴定 A:RT-PCR鉴定新外显子和内含子滞留的引物位置(箭头所示)示意图。由上到下分别鉴定E76、E85、E56、I1、I5和I7;虚线框代表引物预计扩增出的片段。B:RT-PCR鉴定新外显子和内含子滞留的琼脂糖凝胶电泳分析。M:DL2000 DNA marker;泳道1~6依次为RT-PCR扩增E76、E85、E56、I1、I5和I7的琼脂糖电泳图,在鉴定I5的电泳图中分子量大的条带为cNRG4-a,分子量小的条带为cNRG4-f,两者均用红色方框标记。C:cNRG4-f和cNRG4基因mRNA序列(NM_001030544.4)外显子分析结果。E5序列长度为150 bp,E5?序列长度71 bp。"
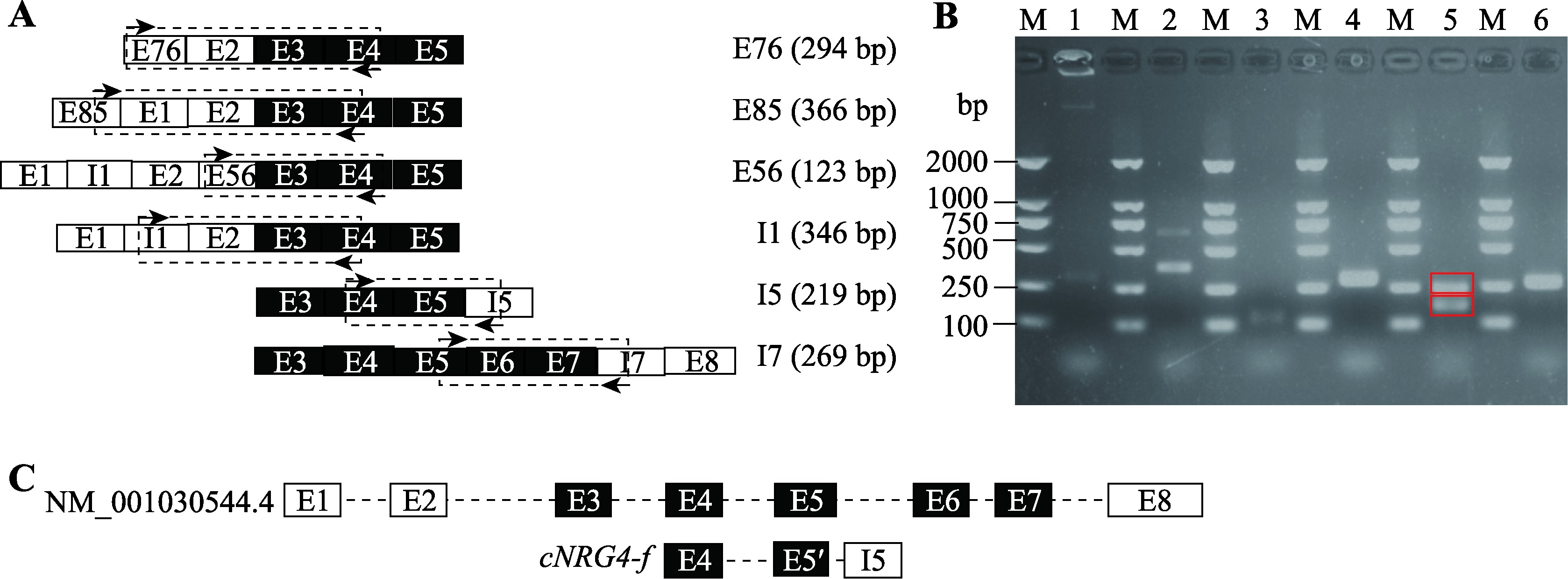
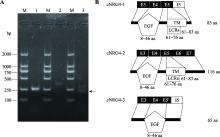
图5
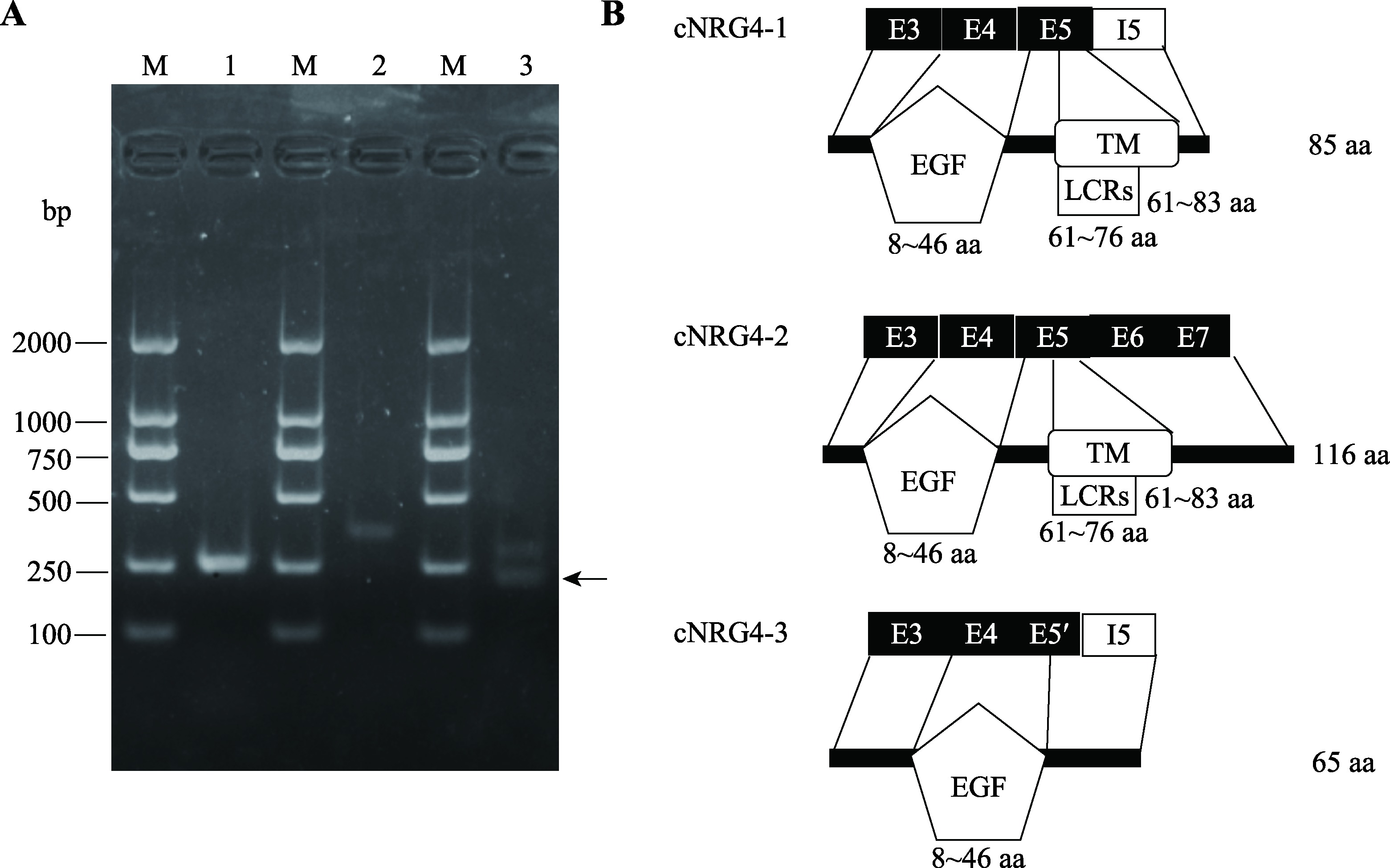
鸡NRG4蛋白质异构体的鉴定与分析 A:cNRG4基因3种不同CDS的 RT-PCR鉴定分析。M:DL2000 DNA marker;泳道1:CDS1的RT-PCR扩增片段;泳道2:CDS2的RT-PCR扩增片段;泳道3:CDS3的RT-PCR扩增片段。箭头所指的条带为目标条带。B:cNRG4蛋白异构体的结构域分析。LCRs:low-complexity regions,cNRG4-1全长有85个氨基酸(aa),其中8~46 aa为EGF结构域,61~83 aa为跨膜结构域(TM),61~76 aa为低复杂性区域(LCRs)。cNRG4-2全长116 aa,其中8~46 aa为EGF结构域,61~83 aa为跨膜结构域(TM),61~76 aa为低复杂性区域(LCRs)。cNRG4-3全长65 aa,其中8~46 aa为EGF结构域。"
| [1] |
Yang F, Li XN. Research progress of neuregulin 4 biological function. Acta Physiol Sin, 2017, 69(3): 351-356.
pmid: 28638929 |
|
杨帆, 李晓南. 神经调节蛋白4生物学功能的研究进展. 生理学报, 2017, 69(3): 351-356.
pmid: 28638929 |
|
| [2] |
Ledonne A, Mercuri NB. On the modulatory roles of neuregulins/ErbB signaling on synaptic plasticity. Int J Mol Sci, 2019, 21(1): 275.
doi: 10.3390/ijms21010275 |
| [3] |
Dai YN, Zhu JZ, Fang ZY, Zhao DJ, Wan XY, Zhu HT, Yu CH, Li YM. A case-control study: association between serum neuregulin 4 level and non-alcoholic fatty liver disease. Metabolism, 2015, 64(12): 1667-1673.
doi: 10.1016/j.metabol.2015.08.013 |
| [4] |
Wang GX, Zhao XY, Meng ZX, Kern M, Dietrich A, Chen ZM, Cozacov Z, Zhou DQ, Okunade AL, Su X, Li SM, Blüher M, Lin JD. The brown fat-enriched secreted factor Nrg4 preserves metabolic homeostasis through attenuation of hepatic lipogenesis. Nat Med, 2014, 20(12): 1436-1443.
doi: 10.1038/nm.3713 |
| [5] |
Chen LL, Peng MM, Zhang JY, Hu X, Min J, Huang QL, Wan LM. Elevated circulating Neuregulin4 level in patients with diabetes. Diabetes Metab Res Rev, 2017, 33(4): e2870.
doi: 10.1002/dmrr.2870 |
| [6] |
Steinthorsdottir V, Stefansson H, Ghosh S, Birgisdottir B, Bjornsdottir S, Fasquel AC, Olafsson O, Stefansson K, Gulcher JR.Multiple novel transcription initiation sites for NRG1. Gene, 2004, 342(1): 97-105.
pmid: 15527969 |
| [7] |
Rimer M, Prieto AL, Weber JL, Colasante C, Ponomareva O, Fromm L, Schwab MH, Lai C, Burden SJ.Neuregulin-2 is synthesized by motor neurons and terminal Schwann cells and activates acetylcholine receptor transcription in muscle cells expressing ErbB4. Mol Cell Neurosci, 2004, 26(2): 271-281.
pmid: 15207852 |
| [8] |
Hayes NVL, Gullick WJ. The neuregulin family of genes and their multiple splice variants in breast cancer. J Mammary Gland Biol Neoplasia, 2008, 13(2): 205-214.
doi: 10.1007/s10911-008-9078-4 |
| [9] |
Carteron C, Ferrer-Montiel A, Cabedo H. Characterization of a neural-specific splicing form of the human neuregulin 3 gene involved in oligodendrocyte survival. J Cell Sci, 2006, 119(Pt 5): 898-909.
doi: 10.1242/jcs.02799 pmid: 16478787 |
| [10] |
Hayes NVL, Newsam RJ, Baines AJ, Gullick WJ. Characterization of the cell membrane-associated products of the neuregulin 4 gene. Oncogene, 2008, 27(5): 715-720.
pmid: 17684490 |
| [11] |
Hayes NVL, Blackburn E, Smart LV, Boyle MM, Russell GA, Frost TM, Morgan BJT, Baines AJ, Gullick WJ. Identification and characterization of novel spliced variants of neuregulin 4 in prostate cancer. Clin Cancer Res, 2007, 13(11): 3147-3155.
doi: 10.1158/1078-0432.CCR-06-2237 pmid: 17545517 |
| [12] | Guo YQ, Wang WJ, Gao ZH, Mu F, Xu HD, Li H, Wang N. Cloning, expression and promoter analysis of adipokine NRG4 gene in chicken. Chin J Agric Biotechol, 2021, 29(11): 2129-2138. |
| 郭亚琦, 王伟佳, 高智慧, 牟芳, 徐海冬, 李辉, 王宁. 鸡脂肪细胞因子NRG4基因的克隆、表达及启动子分析. 农业生物技术学报, 2021, 29(11): 2129-2138. | |
| [13] |
Pfeifer A. NRG4: an endocrine link between brown adipose tissue and liver. Cell Metab, 2015, 21(1): 13-14.
doi: 10.1016/j.cmet.2014.12.008 pmid: 25565202 |
| [14] | Haberle V, Stark A. Eukaryotic core promoters and the functional basis of transcription initiation. Nat Rev Mol Cell Biol, 2018, 19(10): 621-637. |
| [15] |
Forutan M, Ross E, Chamberlain AJ, Nguyen L, Mason B, Moore S, Garner JB, Xiang RD, Hayes BJ. Evolution of tissue and developmental specificity of transcription start sites in Bos taurus indicus. Commun Biol, 2021, 4(1): 829.
doi: 10.1038/s42003-021-02340-6 pmid: 34211114 |
| [16] |
Mejía-Guerra MK, Li W, Galeano NF, Vidal M, Gray J, Doseff AI, Grotewold E. Core promoter plasticity between maize tissues and genotypes contrasts with predominance of sharp transcription initiation sites. Plant Cell, 2015, 27(12): 3309-3320.
doi: 10.1105/tpc.15.00630 |
| [17] |
Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CAM, Taylor MS, Engström PG, Frith MC, Forrest ARR, Alkema WB, Tan SL, Plessy C, Kodzius R, Ravasi T, Kasukawa T, Fukuda S, Kanamori-Katayama M, Kitazume Y, Kawaji H, Kai C, Nakamura M, Konno H, Nakano K, Mottagui-Tabar S, Arner P, Chesi A, Gustincich S, Persichetti F, Suzuki H, Grimmond SM, Wells CA, Orlando V, Wahlestedt C, Liu ET, Harbers M, Kawai J, Bajic VB, Hume DA, Hayashizaki Y. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet, 2006, 38(6): 626-635.
doi: 10.1038/ng1789 pmid: 16645617 |
| [18] |
Thieffry A, López-Márquez D, Bornholdt J, Malekroudi MG, Bressendorff S, Barghetti A, Sandelin A, Brodersen P. PAMP-triggered genetic reprogramming involves widespread alternative transcription initiation and an immediate transcription factor wave. Plant Cell, 2022, 34(7): 2615-2637.
doi: 10.1093/plcell/koac108 |
| [19] |
Braunschweig U, Barbosa-Morais NL, Pan Q, Nachman EN, Alipanahi B, Gonatopoulos-Pournatzis T, Frey B, Irimia M, Blencowe BJ. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res, 2014, 24(11): 1774-1786.
doi: 10.1101/gr.177790.114 pmid: 25258385 |
| [20] |
Hammarskjöld ML. Regulation of retroviral RNA export. Semin Cell Dev Biol, 1997, 8(1): 83-90.
pmid: 15001110 |
| [21] |
Ner-Gaon H, Halachmi R, Savaldi-Goldstein S, Rubin E, Ophir R, Fluhr R. Intron retention is a major phenomenon in alternative splicing in arabidopsis. Plant J, 2004, 39(6): 877-885.
doi: 10.1111/j.1365-313X.2004.02172.x pmid: 15341630 |
| [22] | Rekosh D, Hammarskjold ML. Intron retention in viruses and cellular genes: detention, border controls and passports. Wiley Interdiscip Rev RNA, 2018, 9(3): e1470. |
| [23] |
Marquez Y, Höpfler M, Ayatollahi Z, Barta A, Kalyna M. Unmasking alternative splicing inside protein-coding exons defines exitrons and their role in proteome plasticity. Genome Res, 2015, 25(7): 995-1007.
doi: 10.1101/gr.186585.114 pmid: 25934563 |
| [24] |
Tahmasebi S, Jafarnejad SM, Tam IS, Gonatopoulos- Pournatzis T, Matta-Camacho E, Tsukumo Y, Yanagiya A, Li WC, Atlasi Y, Caron M, Braunschweig U, Pearl D, Khoutorsky A, Gkogkas CG, Nadon R, Bourque G, Yang XJ, Tian B, Stunnenberg HG, Yamanaka Y, Blencowe BJ, Giguère V, Sonenberg N.Control of embryonic stem cell self-renewal and differentiation via coordinated alternative splicing and translation of YY2. Proc Natl Acad Sci USA, 2016, 113(44): 12360-12367.
pmid: 27791185 |
| [25] |
Weatheritt RJ, Sterne-Weiler T, Blencowe BJ. The ribosome-engaged landscape of alternative splicing. Nat Struct Mol Biol, 2016, 23(12): 1117-1123.
doi: 10.1038/nsmb.3317 pmid: 27820807 |
| [26] |
Sun SY, Zhang Z, Sinha R, Karni R, Krainer AR. SF2/ASF autoregulation involves multiple layers of post- transcriptional and translational control. Nat Struct Mol Biol, 2010, 17(3): 306-312.
doi: 10.1038/nsmb.1750 |
| [27] |
Thiele A, Nagamine Y, Hauschildt S, Clevers H. AU-rich elements and alternative splicing in the beta-catenin 3'UTR can influence the human beta-catenin mRNA stability. Exp Cell Res, 2006, 312(12): 2367-2378.
pmid: 16696969 |
| [28] |
Nourse J, Spada S, Danckwardt S. Emerging roles of RNA 3'-end cleavage and polyadenylation in pathogenesis, diagnosis and therapy of human disorders. Biomolecules, 2020, 10(6): 915.
doi: 10.3390/biom10060915 |
| [29] |
Chen W, Jia Q, Song YF, Fu HH, Wei G, Ni T. Alternative polyadenylation: methods, findings, and impacts. Genomics Proteomics Bioinformatics, 2017, 15(5): 287-300.
doi: 10.1016/j.gpb.2017.06.001 |
| [30] |
Jambhekar A, Derisi JL. Cis-acting determinants of asymmetric, cytoplasmic RNA transport. RNA, 2007, 13(5): 625-642.
pmid: 17449729 |
| [31] |
Tian B, Manley JL. Alternative polyadenylation of mRNA precursors. Nat Rev Mol Cell Biol, 2017, 18(1): 18-30.
doi: 10.1038/nrm.2016.116 |
| [32] |
Lau JS, Yip CW, Law KM, Leung FC. Cloning and characterization of chicken growth hormone binding protein (cGHBP). Domest Anim Endocrinol, 2007, 33(1): 107-121.
doi: 10.1016/j.domaniend.2006.04.012 |
| [33] |
Ning BL, Huang JX, Xu HD, Lou YQ, Wang WS, Mu F, Yan XH, Li H, Wang N. Genomic organization, intragenic tandem duplication, and expression analysis of chicken TGFBR2 gene. Poult Sci, 2022, 101(12): 102169.
doi: 10.1016/j.psj.2022.102169 |
| [34] |
Wu QS, Wright M, Gogol MM, Bradford WD, Zhang N, Bazzini AA. Translation of small downstream ORFs enhances translation of canonical main open reading frames. EMBO J, 2020, 39(17): e104763.
doi: 10.15252/embj.2020104763 |
| [35] |
Bazzini AA, Johnstone TG, Christiano R, Mackowiak SD, Obermayer B, Fleming ES, Vejnar CE, Lee MT, Rajewsky N, Walther TC, Giraldez AJ. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J, 2014, 33(9): 981-993.
doi: 10.1002/embj.201488411 pmid: 24705786 |
| [36] |
Dodbele S, Wilusz JE. Ending on a high note: downstream ORFs enhance mRNA translational output. EMBO J, 2020, 39(17): e105959.
doi: 10.15252/embj.2020105959 |
| [1] | 张德洋, 周文川, 李佳乐, 王哲鹏. ABCG2基因错义突变C21R与略阳乌鸡褐壳显著关联[J]. 遗传, 2024, 46(12): 1066-1075. |
| [2] | 邹娴, 何燕华, 何静怡, 王艳, 舒鼎铭, 罗成龙. 鸡原始生殖细胞转染条件优化[J]. 遗传, 2021, 43(3): 280-288. |
| [3] | 王冰源, 牟玉莲, 李奎, 刘志国. 农业动物干细胞研究进展[J]. 遗传, 2020, 42(11): 1073-1080. |
| [4] | 冷奇颖, 郑嘉辉, 徐海冬, PatriciaAdu-Asiamah, 张颖, 杜炳旺, 张丽. 鸡胰岛素降解酶基因环状转录本克隆及其表达规律[J]. 遗传, 2019, 41(12): 1129-1137. |
| [5] | 陈家辉, 任学义, 李丽敏, 卢诗意, 程湉, 谭量天, 梁少东, 何丹林, 罗庆斌, 聂庆华, 张细权, 罗文. 转录组测序揭示细胞周期通路参与鸡腹脂沉积[J]. 遗传, 2019, 41(10): 962-973. |
| [6] | 王凤红,张磊,李晓凯,范一星,乔贤,龚高,严晓春,张令天,王志英,王瑞军,刘志红,王志新,何利兵,张燕军,李金泉,赵艳红,苏蕊. 山羊基因组研究进展[J]. 遗传, 2019, 41(10): 928-938. |
| [7] | 褚衍凯,靳艳飞,邢天宇,马广伟,崔婷婷,闫晓红,李辉,王宁. 鸡PPARγ基因转录本3上游开放阅读框转录后的调控作用[J]. 遗传, 2018, 40(8): 657-667. |
| [8] | 李敏,董翔宇,梁浩,冷丽,张慧,王守志,李辉,杜志强. 肉鸡腹脂率双向选择系群体表型数据库(NEAUHLFPD)的设计及其功能实现[J]. 遗传, 2017, 39(5): 430-437. |
| [9] | 张潇飞,宋鹤,刘静,张文建,闫晓红,李辉,王宁. 鸡miR-17-92基因簇靶基因ZFPM2的鉴定及功能分析[J]. 遗传, 2017, 39(4): 333-345. |
| [10] | 李光奇, 孙从佼, 吴桂琴, 石凤英, 刘爱巧, 孙皓, 杨宁. 利用转录组测序筛选鸡蛋褐壳性状相关基因[J]. 遗传, 2017, 39(11): 1102-1111. |
| [11] | 程敏, 张文建, 邢天宇, 闫晓红, 李玉茂, 李辉, 王宁. 鸡miR-17-92基因簇上游调控区功能分析[J]. 遗传, 2016, 38(8): 724-735. |
| [12] | 杨盛智, 吴国艳, 龙梅, 邓雯文, 王红宁, 邹立扣. 鸡蛋生产链中沙门氏菌对抗生素及消毒剂的耐药性研究[J]. 遗传, 2016, 38(10): 948-956. |
| [13] | 张涛, 王文浩, 张跟喜, 王金玉, 薛倩, 顾玉萍. 京海黄鸡体重性状全基因组关联分析[J]. 遗传, 2015, 37(8): 811-820. |
| [14] | 马俊平, 杨犀, 律娜, 刘飞, 陈燕, 朱宝利. 鸡T细胞受体γ链基因位点重新测序和组装[J]. 遗传, 2015, 37(6): 568-574. |
| [15] | 宋晓燕, 张德祥, 张文武, 季从亮, 张细权, 罗庆斌. 鸡3种组织中热应激相关基因的表达谱芯片分析[J]. 遗传, 2014, 36(8): 800-808. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||

www.chinagene.cn
备案号:京ICP备09063187号-4
总访问:,今日访问:,当前在线:
