遗传 ›› 2024, Vol. 46 ›› Issue (8): 649-660.doi: 10.16288/j.yczz.24-035
闵羽1,2( ), 倪子涵1,2, 马玲玲1,2, 渡边嘉典1,2(
), 倪子涵1,2, 马玲玲1,2, 渡边嘉典1,2( )
)
收稿日期:2024-03-08
修回日期:2024-05-07
出版日期:2024-08-20
发布日期:2024-05-14
通讯作者:
渡边嘉典,教授,研究方向:染色体分离机制。E-mail: ywatanabe@jiangnan.edu.cn作者简介:闵羽,硕士研究生,专业方向:生物与医药。E-mail: 1401577396@qq.com
Yu Min1,2( ), Zihan Ni1,2, Lingling Ma1,2, Yoshinori Watanabe1,2(
), Zihan Ni1,2, Lingling Ma1,2, Yoshinori Watanabe1,2( )
)
Received:2024-03-08
Revised:2024-05-07
Published:2024-08-20
Online:2024-05-14
摘要:
减数分裂特异性调控分子Moa1定位到着丝粒受到动粒蛋白CENP-C的调控,同时Moa1参与黏连蛋白Rec8介导的着丝粒区域姐妹染色单体的黏连。为了研究这些蛋白质之间的相互作用,本研究利用酵母双杂交实验(yeast two-hybrid assay)测定分析了Moa1和CENP-C、Rec8之间的相互作用,并通过在Moa1中定点突变鉴定了与CENP-C和Rec8相互作用所需的一些氨基酸残基。实验结果表明,Moa1和CENP-C的相互作用对于Moa1参与调节姐妹动粒的单极附着很重要。然而,双杂交实验中与Rec8相互作用所需的Moa1的S143和T150突变没有显示出Moa1或Rec8功能的显著缺陷。这表明氨基酸残基的突变可能不足以干扰体内Moa1和Rec8之间的相互作用,需要进一步的研究来确定Moa1和Rec8的相互作用域。本研究揭示了影响减数分裂同源染色体分离的Moa1氨基酸位点,为减数分裂的染色体分离机制提供更深入的理解。
闵羽, 倪子涵, 马玲玲, 渡边嘉典. Moa1与CENP-C和Rec8的相互作用及其在裂殖酵母减数分裂中的功能[J]. 遗传, 2024, 46(8): 649-660.
Yu Min, Zihan Ni, Lingling Ma, Yoshinori Watanabe. Functional roles of the interaction of Moa1 with CENP-C and Rec8 in meiosis of Schizosaccharomyces pombe[J]. Hereditas(Beijing), 2024, 46(8): 649-660.
表1
本研究所用菌株及其基因型"
| 菌株名称 | 基因型 |
|---|---|
| PW632 | h+ pat1-114 3pk-moa1 |
| PW1 | h+ pat1-114 natMX6-3pk-moa1 |
| M1 | h- leu1 imr1L-GFP mes1-B44 rec8-2A<<Kan rec12::hyg |
| M2 | h+ mes1-B44 rec8-2A<<Kan rec12::hyg |
| M3 | h90 mei4::hyg |
| PM11 | h- leu1 imr1L-GFP mes1-B44 rec8-2A<<Kan rec12::hyg natMX6-3pk-moa1-S143K T150K |
| PM12 | h+ mes1-B44 rec8-2A<<Kan rec12::hyg natMX6-3pk-moa1-S143K T150K |
| PM21 | h- leu1 imr1L-GFP mes1-B44 rec8-2A<<Kan rec12::hyg natMX6-3pk-moa1-S143D T150E |
| PM22 | h+ mes1-B44 rec8-2A<<Kan rec12::hyg natMX6-3pk-moa1-S143D T150E |
| PM31 | h- leu1 imr1L-GFP mes1-B44 rec8-2A<<Kan rec12::hyg natMX6-3pk-moa1-E165K |
| PM32 | h+ mes1-B44 rec8-2A<<Kan rec12::hyg natMX6-3pk-moa1-E165K |
| PM41 | h- leu1 imr1L-GFP mes1-B44 rec8-2A<<Kan rec12::hyg natMX6-3pk-moa1-D167K E168K |
| PM42 | h+ mes1-B44 rec8-2A<<Kan rec12::hyg natMX6-3pk-moa1-D167K E168K |
| PM51 | h- leu1 imr1L-GFP mes1-B44 rec8-2A<<Kan rec12::hyg natMX6-3pk-moa1-I170K L171K |
| PM52 | h+ mes1-B44 rec8-2A<<Kan rec12::hyg natMX6-3pk-moa1-I170K L171K |
| LM1 | h- leu1imr1L-GFP mes1-B44 rec8-2A<<Kan rec12::bsdr moa1::natr |
| LM2 | h+ mes1-B44 rec8-2A<<Kan rec12::bsdr moa1::natr |
| PM-W | h90 mei4::hyg natMX6-GFP-3pk-moa1 |
| PM36 | h90 mei4::hyg natMX6-GFP-3pk-moa1-E165K |
| PM37 | h90 mei4::hyg natMX6-GFP-3pk-moa1-D167K E168K |
| PM38 | h90 mei4::hyg natMX6-GFP-3pk-moa1-I170K L171K |
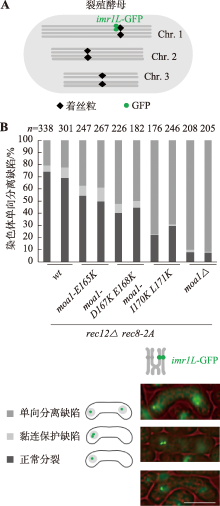
图3
moa1-mr突变体的染色体分离行为 A:裂殖酵母染色体imr1L-GFP示意图。B:在减数分裂I分裂后的细胞中监测一个同源物上标记的imr1L-GFP的分离模式。减数分裂特异蛋白Mes1通过抑制后期促进复合物(anaphase- promoting complex,APC),控制减数第一次分裂到第二次分裂的转变,mes1-B44突变可以使减数分裂细胞停在第二次分裂前期[44,45]。携带imr1L-GFP的杂交细胞因mes1-B44突变停滞在减数第二次分裂前期便于对减数第一次分裂期间的染色体分离情况进行分析。右侧显示了不同情况的染色体分离行为:两边各一个信号点代表姐妹染色单体分离,一边两个信号点代表姐妹染色单体黏连缺陷,一边一个信号点代表染色体单向分离。相同基因型的杂交细胞做独立的两组数据,每一组具有统计意义的细胞数n >100。比例尺:10 μm。"
| [1] | Börner GV, Hochwagen A, Macqueen AJ. Meiosis in budding yeast. Genetics, 2023, 225(2): 1-33. |
| [2] | Hochwagen A. Meiosis. Curr Biol, 2008, 18(15): R641-R645. |
| [3] |
Sakuno T, Watanabe Y. Studies of meiosis disclose distinct roles of cohesion in the core centromere and pericentromeric regions. Chromosome Res, 2009, 17(2): 239-249.
doi: 10.1007/s10577-008-9013-y pmid: 19308704 |
| [4] |
Kagami A, Sakuno T, Yamagishi Y, Ishiguro T, Tsukahara T, Shirahige K, Tanaka K, Watanabe Y. Acetylation regulates monopolar attachment at multiple levels during meiosis I in fission yeast. EMBO Rep, 2011, 12(11): 1189-1195.
doi: 10.1038/embor.2011.188 pmid: 21979813 |
| [5] |
Yokobayashi S, Watanabe Y. The kinetochore protein Moal enables cohesion-mediated monopolar attachment at meiosis I. Cell, 2005, 123(5): 803-817.
pmid: 16325576 |
| [6] |
Lee BH, Kiburz BM, Amon A. Spo13 maintains centromeric cohesion and kinetochore coorientation during meiosis I. Curr Biol, 2004, 14(24): 2168-2182.
pmid: 15620644 |
| [7] | Kim J, Ishiguro K-I, Nambu A, Akiyoshi B, Yokobayashi S, Kagami A, Ishiguro T, Pendas AM, Takeda N, Sakakibara Y, Kitajima TS, Tanno Y, Sakuno T, Watanabe Y. Meikin is a conserved regulator of meiosis-I-specific kinetochore function. Nature, 2014, 517(7535): 466-471. |
| [8] | Hayashi T, Ebe M, Nagao K, Kokubu A, Sajiki K, Yanagida M. Schizosaccharomyces pombe centromere protein Mis19 links Mis16 and Mis18 to recruit CENP-A through interacting with NMD factors and the SWI/SNF complex. Genes Cells, 2014, 19(7): 541-554. |
| [9] |
Earnshaw WC, Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma, 1985, 91(3-4): 313-321.
doi: 10.1007/BF00328227 pmid: 2579778 |
| [10] | She CW, Song YC. Advances in research of the structure and function of plant centromeres. Hereditas(Beijing), 2006, 28(12): 1597-1606. |
| 佘朝文, 宋运淳. 植物着丝粒结构和功能的研究进展. 遗传, 2006, 28(12): 1597-1606. | |
| [11] |
Przewloka MR, Venkei Z, Bolanos-Garcia VM, Debski J, Dadlez M, Glover DM. CENP-C is a structural platform for kinetochore assembly. Curr Biol, 2011, 21(5): 399-405.
doi: 10.1016/j.cub.2011.02.005 pmid: 21353555 |
| [12] | Hara M, Ariyoshi M, Sano T, Nozawa RS, Shinkai S, Onami S, Jansen I, Hirota T, Fukagawa T. Centromere/ kinetochore is assembled through CENP-C oligomerization. Mol Cell, 2023, 83(13): 2188-2205.e13. |
| [13] | Chik JK, Moiseeva V, Goel PK, Meinen BA, Koldewey P, An SJ, Mellone BG, Subramanian L, Cho US. Structures of CENP-C cupin domains at regional centromeres reveal unique patterns of dimerization and recruitment functions for the inner pocket. J Biol Chem, 2019, 294(38): 14119-14134. |
| [14] | Musacchio A, Desai A. A molecular view of kinetochore assembly and function. Biology (Basel), 2017, 6(1): 5. |
| [15] | Kwon M-S, Hori T, Okada M, Fukagawa T. CENP-C is involved in chromosome segregation, mitotic checkpoint function, and kinetochore assembly. Mol Biol Cell, 2007, 18(6): 2155-2168. |
| [16] | Fellmeth JE, Jang JK, Persaud M, Sturm H, Changela N, Parikh A, Mckim KS. A dynamic population of prophase CENP-C is required for meiotic chromosome segregation. PLoS Genet, 2023, 19(11): e1011066. |
| [17] | Heeger S, Leismann O, Schittenhelm R, Schraidt O, Heidmann S, Lehner CF. Genetic interactions of separase regulatory subunits reveal the diverged Drosophila Cenp-C homolog. Genes Dev, 2005, 19(17): 2041-2053. |
| [18] |
Moore LL, Roth MB. Hcp-4, a Cenp-C-like protein in Caenorhabditis elegans, is required for resolution of sister centromeres. J Cell Biol, 2001, 153(6): 1199-1208.
doi: 10.1083/jcb.153.6.1199 pmid: 11402064 |
| [19] |
Fellmeth JE, Mckim KS. Meiotic CENP-C is a shepherd: bridging the space between the centromere and the kinetochore in time and space. Essays Biochem, 2020, 64(2): 251-261.
doi: 10.1042/EBC20190080 pmid: 32794572 |
| [20] |
Hillers KJ, Jantsch V, Martinez-Perez E, Yanowitz JL. Meiosis. WormBook, 2017, 2017: 1-43.
doi: 10.1895/wormbook.1.178.1 pmid: 26694509 |
| [21] | XU XY, Yanagida M. Cohesin organization, dynamics, and subdomain functions revealed by genetic suppressor screening. Proc Jpn Acad Ser B Phys Biol Sci, 2023, 99(3): 61-74. |
| [22] | Zhang N, Zhang J, Lin G. Advances in the study of DNA damage and repair in mammalian oocytes. Hereditas (Beijing), 2023, 45(5): 379-394. |
| 张楠, 张珏, 林戈. 哺乳动物卵母细胞的DNA损伤与修复研究进展. 遗传, 2023, 45(5): 379-394. | |
| [23] | Kuru-Schors M, Haemmerle M, Gutschner T. The cohesin complex and its interplay with non-coding RNAs. Noncoding RNA, 2021, 7(4): 67. |
| [24] | Litwin I, Pilarczyk E, Wysocki R. The emerging role of cohesin in the DNA damage response. Genes (Basel), 2018, 9(12): 581. |
| [25] | Zhang Y, Fang YD. Progresses on the structure and function of cohesin. Hereditas (Beijing), 2020, 42(1): 57-72. |
| 张雨, 方玉达. Cohesin结构及功能研究进展. 遗传, 2020, 42(1): 57-72. | |
| [26] |
Yokobayashi S, Yamamoto M, Watanabe Y. Cohesins determine the attachment manner of kinetochores to spindle microtubules at meiosis I in fission yeast. Mol Cell Biol, 2003, 23(11): 3965-3973.
doi: 10.1128/MCB.23.11.3965-3973.2003 pmid: 12748297 |
| [27] | Rittenhouse NL, Dowen JM. Cohesin regulation and roles in chromosome structure and function. Curr Opin Genet Dev, 2024, 85: 102159. |
| [28] | Lu YJ, Zhou CY, Xiong B. Functional diversity of chromosome cohesion proteins. SCI SIN Vitae, 2022, 52(12): 1844-1857. |
| 卢亚娟, 周长银, 熊波. 染色体黏合蛋白功能的多样性. 中国科学:生命科学, 2022, 52(12): 1844-1857. | |
| [29] | Minamino M, Higashi TL, Bouchoux C, Uhlmann F. Topological in vitro loading of the budding yeast cohesin ring onto DNA. Life Sci Alliance, 2018, 1(5): e201800143. |
| [30] |
Kitajima TS, Miyazaki Y, Yamamoto M, Watanabe Y. Rec8 cleavage by separase is required for meiotic nuclear divisions in fission yeast. EMBO J, 2003, 22(20): 5643-5653.
pmid: 14532136 |
| [31] |
Takahashi TS, Yiu PY, Chou MF, Gygi S, Walter JC. Recruitment of xenopus Scc2 and cohesin to chromatin requires the pre-replication complex. Nat Cell Biol, 2004, 6(10): 991-996.
doi: 10.1038/ncb1177 pmid: 15448702 |
| [32] |
Parisi S, Mckay MJ, Molnar M, Thompson MA, van der Spek PJ, van Drunen-Schoenmaker E, Kanaar R, Lehmann E, Hoeijmakers JH, Kohli J. Rec8p, a meiotic recombination and sister chromatid cohesion phosphoprotein of the Rad21p family conserved from fission yeast to humans. Mol Cell Biol, 1999, 19(5): 3515-3528.
doi: 10.1128/MCB.19.5.3515 pmid: 10207075 |
| [33] | Kitajima TS, Kawashima SA, Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature, 2004, 427(6974): 510-517. |
| [34] | Watanabe Y, Nurse P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature, 1999, 400(6743): 461-464. |
| [35] |
Miyazaki S, Kim J, Yamagishi Y, Ishiguro T, Okada Y, Tanno Y, Sakuno T, Watanabe Y. Meikin-associated polo-like kinase specifies Bub1 distribution in meiosis I. Genes Cells, 2017, 22(6): 552-567.
doi: 10.1111/gtc.12496 pmid: 28497540 |
| [36] |
Tanaka K, Li Chang H, Kagami A, Watanabe Y. CENP-C functions as a scaffold for effectors with essential kinetochore functions in mitosis and meiosis. Dev Cell, 2009, 17(3): 334-343.
doi: 10.1016/j.devcel.2009.08.004 pmid: 19758558 |
| [37] | Ishiguro KI, Shimada R. MEIOSIN directs initiation of meiosis and subsequent meiotic prophase program during spermatogenesis. Genes Genet Syst, 2022, 97(1): 27-39. |
| [38] | Nimmo ER, Pidoux AL, Perry PE, Allshire RC. Defective meiosis in telomere-silencing mutants of Schizosaccharomyces pombe. Nature, 1998, 392(6678): 825-828. |
| [39] | Ding DQ, Haraguchi T, Hiraoka Y. Chromosomally- retained RNA mediates homologous pairing. Nucleus, 2012, 3(6): 516-519. |
| [40] |
Menees TM, Ross-Macdonald PB, Roeder GS. mei4, a meiosis-specific yeast gene required for chromosome synapsis. Mol Cell Biol, 1992, 12(3): 1340-1351.
doi: 10.1128/mcb.12.3.1340-1351.1992 pmid: 1545815 |
| [41] | Kumar R, Bourbon HM, De Massy B. Functional conservation of Mei4 for meiotic DNA double-strand break formation from yeasts to mice. Genes Dev, 2010, 24(12): 1266-1280. |
| [42] |
Zhou Z, Sang Q, Wang L. Physiological and pathological mechanisms of oocyte meiosis. Hereditas(Beijing), 2023, 45(12): 1087-1099.
doi: 10.16288/j.yczz.23-170 pmid: 38764273 |
| 周舟, 桑庆, 王磊. 人类卵母细胞减数分裂的生理和病理机制. 遗传, 2023, 45(12): 1087-1099. | |
| [43] |
Takahashi K, Murakami S, Chikashige Y, Funabiki H, Niwa O, Yanagida M. A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol Biol Cell, 1992, 3(7): 819-835.
pmid: 1515677 |
| [44] |
Kimata Y, Kitamura K, Fenner N, Yamano H. Mes1 controls the meiosis I to meiosis II transition by distinctly regulating the anaphase-promoting complex/cyclosome coactivators Fzr1/Mfr1 and Slp1 in fission yeast. Mol Biol Cell, 2011, 22(9): 1486-1494.
doi: 10.1091/mbc.E10-09-0774 pmid: 21389117 |
| [45] | Izawa D, Goto M, Yamashita A, Yamano H, Yamamoto M. Fission yeast Mes1p ensures the onset of meiosis II by blocking degradation of cyclin Cdc13p. Nature, 2005, 434(7032): 529-533. |
| [46] |
Ogushi S, Rattani A, Godwin J, Metson J, Schermelleh L, Nasmyth K. Loss of sister kinetochore co-orientation and peri-centromeric cohesin protection after meiosis I depends on cleavage of centromeric REC8. Dev Cell, 2021, 56(22): 3100-3114.e4.
doi: 10.1016/j.devcel.2021.10.017 pmid: 34758289 |
| [47] | Sakuno T, Hiraoka Y. Rec8 cohesin: a structural platform for shaping the meiotic chromosomes. Genes (Basel), 2022, 13(2). 200. |
| [48] | Ma W, Zhou JW, Chen J, Carr AM, Watanabe Y. Meikin synergizes with shugoshin to protect cohesin Rec8 during meiosis I. Genes Dev, 2021, 35(9-10): 692-697. |
| [49] | Mehta G, Anbalagan GK, Bharati AP, Gadre P, Ghosh SK. An interplay between Shugoshin and Spo13 for centromeric cohesin protection and sister kinetochore mono-orientation during meiosis I in Saccharomyces cerevisiae. Curr Genet, 2018, 64(5): 1141-1152. |
| [1] | 倪子涵, 闵羽, 马玲玲, 渡边嘉典. 着丝粒蛋白Fta2磷酸化对减数分裂的影响[J]. 遗传, 2024, 46(7): 552-559. |
| [2] | 闫静亮, 马玲玲, 渡边嘉典. Ssu72磷酸酶缺失导致减数第二次分裂过程中纺锤体交叉[J]. 遗传, 2024, 46(6): 502-508. |
| [3] | 吕香江, 郭静, 林戈. TRIP13基因新突变导致卵母细胞成熟阻滞为特征的女性不孕[J]. 遗传, 2023, 45(6): 514-525. |
| [4] | 周舟, 桑庆, 王磊. 人类卵母细胞减数分裂的生理和病理机制[J]. 遗传, 2023, 45(12): 1087-1099. |
| [5] | 郭雨萱, 严顺平, 王应祥. 重组酶RAD51和DMC1功能保守和分化研究进展[J]. 遗传, 2022, 44(5): 398-413. |
| [6] | 李园园, 郭磊, 韩之明. NEK家族在细胞周期调控中的作用[J]. 遗传, 2021, 43(7): 642-653. |
| [7] | 聂辉, 张译文, 李佳宁, 王楠楠, 徐澜. 减数分裂联会复合体异常与不孕不育相关性研究进展[J]. 遗传, 2021, 43(12): 1142-1148. |
| [8] | 李帆, 余蓉培, 孙丹, 王继华, 李绅崇, 阮继伟, 单芹丽, 陆平利, 汪国鲜. 抑制植物减数分裂重组的分子机理[J]. 遗传, 2019, 41(1): 52-65. |
| [9] | 廖亚平,王春景,梁猛,胡小梅,吴琦. 平衡复杂染色体重排携带者的遗传与生育情况分析[J]. 遗传, 2017, 39(5): 396-412. |
| [10] | 岳珊珊,夏来新. 酵母双杂交筛选与果蝇C(2)M相互作用的蛋白[J]. 遗传, 2015, 37(11): 1160-1166. |
| [11] | 张宝乐 高殿帅 徐银学. G蛋白偶联受体3:调控神经系统和卵泡发育的关键因子[J]. 遗传, 2013, 35(5): 578-586. |
| [12] | 谢文军,史典义,蔡泽熙,陈晓阳,金危危. 联会复合体的组成、功能及遗传控制[J]. 遗传, 2012, 34(2): 167-176. |
| [13] | 刘梦豪,赵凯强,王雅栋,杨梦平,赵宁宁,杨大祥. 蝗虫精母细胞减数分裂各时期的识别[J]. 遗传, 2012, 34(12): 1628-1637. |
| [14] | 段涛,杨庆岭,王刘,史庆华,于德新. 人精母细胞重组频率和年龄相关性的分析[J]. 遗传, 2011, 33(7): 725-730. |
| [15] | 陈军,罗伟雄,李明,罗琼. 水稻减数分裂过程中染色体重组交换行为[J]. 遗传, 2011, 33(6): 648-653. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||

www.chinagene.cn
备案号:京ICP备09063187号-4
总访问:,今日访问:,当前在线:
