遗传 ›› 2024, Vol. 46 ›› Issue (8): 589-602.doi: 10.16288/j.yczz.24-149
收稿日期:2024-05-24
修回日期:2024-07-06
出版日期:2024-08-20
发布日期:2024-07-10
通讯作者:
李青,博士,副教授,研究方向:细胞生物学。E-mail: qing_napier@126.com;作者简介:王纪龙,硕士研究生,专业方向:病原生物学。E-mail: wjlshuaifeng@163.com
基金资助:
Jilong Wang1( ), Qing Li2,3(
), Qing Li2,3( ), Tingzheng Zhan1,3(
), Tingzheng Zhan1,3( )
)
Received:2024-05-24
Revised:2024-07-06
Published:2024-08-20
Online:2024-07-10
Supported by:摘要:
自转录活性调节区测序(self-transcribing active regulatory region sequencing,STARR-seq)是一种可发现并同时验证全基因组增强子活性的高通量测序方法。其原理为:将待验证序列插入质粒载体并电转入细胞中,该序列在作为增强子提高靶基因转录的同时,其本身也作为靶基因被增强转录。通过对转录组进行测序,并对比未插入片段的测序结果,可获得增强子在基因组位置及活性的信息。在传统增强子研究方法中,通过对染色质开放区域和转录活性区域进行测序以预测增强子,但只能逐一验证预测结果,无法高通量验证增强子活性。STARR-seq技术解决了上述缺陷,可在对全基因组增强子高通量挖掘的同时,对其活性进行可靠的验证。自STARR-seq技术发明以来,已被广泛运用于不同物种与细胞中的增强子发现及活性验证研究。本文对传统增强子预测方法以及STARR-seq技术的基本原理、发展历史和具体运用进行了介绍,并对其发展前景进行展望,以期为后续增强子相关领域研究人员提供参考。
王纪龙, 李青, 战廷正. 自转录活性调节区测序技术在增强子发现研究中的应用[J]. 遗传, 2024, 46(8): 589-602.
Jilong Wang, Qing Li, Tingzheng Zhan. Principle and application of self-transcribing active regulatory region sequencing in enhancer discovery research[J]. Hereditas(Beijing), 2024, 46(8): 589-602.
表1
传统增强子研究方法的对比"
| 基本原理 | 方法名称 | 具体方法 | 运用 | 优点 | 缺点 | 参考文献 |
|---|---|---|---|---|---|---|
| 检测DNA可及区或染色质开放区域 | DNase-seq | 通过识别DNase I 超敏位点判断顺式调控元件所在区域后预测增强子 | 染色质开放区域检测 | 高通量检测全基因组染色质开放区域 | 切割偏好度过大,只能预测增强子 | [ |
| ATAC-seq | 使用链接了测序接头的Tn5转座酶切割染色质获得开放区域文库后进行测序 | 染色质开放区域检测 | 高通量,样本需求量小 | 转座酶切割可能有偏好,只能预测增强子 | [ | |
| MNase-seq | 利用微球核酸酶消化核小体之间裸露区域,富集与核小体结合的DNA片段后测序,绘制核小体位置图谱间接反应染色质开放区域 | 绘制基因组中核小体位置图谱 | 分辨率极高可分析单个核小体 | 间接检测染色质开放区域,工作量大 | [ | |
| FAIRE-seq | 使用甲醛交联染色质互作部位后通过超声进行破碎,通过酚-氯仿抽提分离DNA进行测序 | 染色质开放区域检测 | 使用超声破碎,避免酶切偏好性 | 分辨率较低,背景噪音高 | [ | |
| 检测增强子相关组蛋白修饰 | ChIP-seq | 通过染色质免疫共沉淀特异性富集增强子相关蛋白结合的DNA序列 | 识别转录因子结合位点以及特异性组蛋白修饰 | 分辨率高,直接识别转录因子作用位点或组蛋白修饰 | 需要特异性抗体,只能预测增强子 | [ |
| CUT&Run | 使用Protein A/G结合的MNase,靶向目的蛋白,并在目的蛋白附近对其互作DNA进行片段化 | 研究组蛋白修饰和蛋白质-DNA相互作用 | 对样本需求量较低,高灵敏度低背景噪音 | 需要特异性抗体 | [ | |
| CUT&Tag | 使用Protein A/G结合的转座酶,靶向目的蛋白,并在目的蛋白结合的DNA区域插入测序接头序列并片段化DNA | 高灵敏度低背景噪音的ChIP-seq | 对样本需求量极低,高灵敏度低背景噪音 | 需要特异性抗体 | [ | |
| 直接检测增强子RNA | GRO-seq | 在体外进行新增强子RNA合成及标记后进行测序 | 检测增强子RNA | 动态实时检测增强子活性,尤其是细胞处于不同环境时 | 无法直接分辨增强子RNA与mRNA | [ |

图3
STARR-seq排除传统技术误差 红色阴影区域为STARR-seq所鉴定增强子。STARR-seq结果与DHS-seq、ChIP-seq(分别使用H3K27ac、H3K4me1抗体)结果相比既有相同(左三红色阴影区)也有差异(左一红色阴影区)。DHS-seq显示左一红色阴影区为染色质关闭的区域,H3K27ac ChIP-seq结果显示无乙酰化修饰转录不活跃,根据传统策略应预测为非增强子区域,但是STARR-seq结果显示该区域具有增强子活性。这是因为STARR-seq技术中用于检测的质粒DNA一直处于开放状态,不受折叠影响,因此排除了根据染色质开放区域以及特异性组蛋白修饰产生的预测误差。"
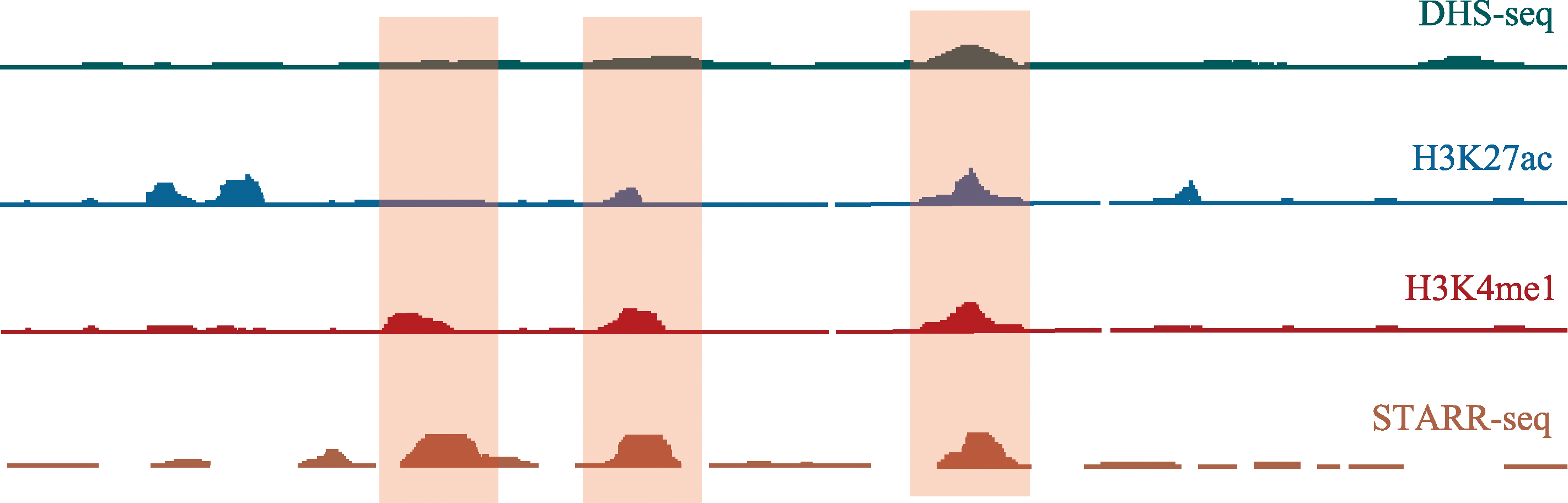
表2
STARR-seq及与其联合使用的各项技术"
| 方法名称 | 具体方法 | 运用 | 优点 | 缺点 | 参考文献 |
|---|---|---|---|---|---|
| STARR-seq | 将待验证片段克隆至核心启动子下游构建待验证文库,将该文库转染至细胞中,培养细胞并提取RNA。活性增强子将提高自我转录程度并成为最终报告转录本的一部分 | 全基因组增强子发掘及活性验证 | 高通量,直接发现并验证全基因组增强子 | 对文库质量以及转染效率有较高要求 | [ |
| AAV-STARR-seq | 通过AAV传递STARR-seq文库后进行测序 | 活体动物体内增强子检测 | 可开展体内增强子检测 | 无法避免染色质状态对增强子活性的影响 | [ |
| ChIP-STARR-seq | 将ChIP-seq鉴定出的DNA连接至STARR-seq载体后进行测序 | 转录因子结合的DNA序列增强子活性检测 | 直接验证转录因子作用位点 | 无法避免染色质状态对增强子活性的影响 | [ |
| ATAC-STARR-seq | 使用ATAC-seq文库连接STARR-seq载体后进行测序 | 准确高效的从染色质开放区域鉴定出增强子 | 可准确检测染色质开放区域增强子 | 无法避免染色质状态对增强子活性的影响 | [ |
| FAIRE-STARR-seq | 使用FAIRE-seq鉴定出的染色质开放区域连接至STARR-seq载体后进行测序 | 准确高效的从染色质开放区域鉴定出增强子 | 可准确检测染色质开放区域增强子 | 无法避免染色质状态对增强子活性的影响 | [ |
| [1] |
Hamamoto K, Fukaya T. Molecular architecture of enhancer-promoter interaction. Curr Opin Cell Biol, 2022, 74: 62-70.
doi: 10.1016/j.ceb.2022.01.003 pmid: 35168174 |
| [2] |
Bianconi E, Piovesan A, Facchin F, Beraudi A, Casadei R, Frabetti F, Vitale L, Pelleri MC, Tassani S, Piva F, Perez-Amodio S, Strippoli P, Canaider S. An estimation of the number of cells in the human body. Ann Hum Biol, 2013, 40(6): 463-471.
doi: 10.3109/03014460.2013.807878 pmid: 23829164 |
| [3] | Han XP, Zhou ZM, Fei LJ, Sun HY, Wang RY, Chen Y, Chen HD, Wang JJ, Tang HN, Ge WH, Zhou YC, Ye F, Jiang MM, Wu JQ, Xiao YY, Jia XN, Zhang TY, Ma XJ, Zhang Q, Bai XL, Lai SJ, Yu CX, Zhu LJ, Lin R, Gao YC, Wang M, Wu YQ, Zhang JM, Zhan RY, Zhu SY, Hu HL, Wang CC, Chen M, Huang H, Liang TB, Chen JH, Wang WL, Zhang D, Guo GJ. Construction of a human cell landscape at single-cell level. Nature, 2020, 581(7808): 303-309. |
| [4] | Venters BJ, Pugh BF. How eukaryotic genes are transcribed. Crit Rev Biochem Mol Biol, 2009, 44(2-3): 117-141. |
| [5] |
Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet, 2009, 10(3): 155-159.
doi: 10.1038/nrg2521 pmid: 19188922 |
| [6] |
Panigrahi A, O'malley BW. Mechanisms of enhancer action: the known and the unknown. Genome Biol, 2021, 22(1): 108.
doi: 10.1186/s13059-021-02322-1 pmid: 33858480 |
| [7] | Zhang Y, See YX, Tergaonkar V, Fullwood MJ. Long- distance repression by human silencers: chromatin interactions and phase separation in silencers. Cells, 2022, 11(9): 1560. |
| [8] |
Devkota P, Wuchty S. Promoter/enhancer-based controllability of regulatory networks. Sci Rep, 2022, 12(1): 3528.
doi: 10.1038/s41598-022-07035-4 pmid: 35241702 |
| [9] | Qi SH, Wang QL, Zhang JY, Liu Q, Li CY. The regulatory mechanisms by enhancers during cancer initiation and progression. Hereditas (Beijing), 2022, 44(4): 275-288. |
| 漆思晗, 王棨临, 张俊有, 刘倩, 李春燕. 增强子调控癌症发生发展的机制研究. 遗传, 2022, 44(4): 275-288. | |
| [10] |
Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell, 2011, 144(3): 327-339.
doi: 10.1016/j.cell.2011.01.024 pmid: 21295696 |
| [11] |
Li GL, Ruan XA, Auerbach RK, Sandhu KS, Zheng MZ, Wang P, Poh HM, Goh Y, Lim J, Zhang JY, Sim HS, Peh SQ, Mulawadi FH, Ong CT, Orlov YL, Hong SZ, Zhang ZZ, Landt S, Raha D, Euskirchen G, Wei CL, Ge WH, Wang HE, Davis C, Fisher-Aylor KI, Mortazavi A, Gerstein M, Gingeras T, Wold B, Sun Y, Fullwood MJ, Cheung E, Liu E, Sung WK, Snyder MK, Ruan YJ. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell, 2012, 148(1-2): 84-98.
doi: 10.1016/j.cell.2011.12.014 pmid: 22265404 |
| [12] |
Kieffer-Kwon KR, Tang ZH, Mathe E, Qian J, Sung MH, Li GL, Resch W, Baek S, Pruett N, Grøntved L, Vian L, Nelson S, Zare H, Hakim O, Reyon D, Yamane A, Nakahashi H, Kovalchuk AL, Zou JZ, Joung JK, Sartorelli V, Wei CL, Ruan XA, Hager GL, Ruan YJ, Casellas R. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell, 2013, 155(7): 1507-1520.
doi: 10.1016/j.cell.2013.11.039 pmid: 24360274 |
| [13] |
Tang ZH, Luo OJ, Li XW, Zheng MZ, Zhu JJ, Szalaj P, Trzaskoma P, Magalska A, Wlodarczyk J, Ruszczycki B, Michalski P, Piecuch E, Wang P, Wang DJ, Tian SZY, Penrad-Mobayed M, Sachs LM, Ruan XA, Wei CL, Liu ET, Wilczynski GM, Plewczynski D, Li GL, Ruan YJ. CTCF-mediated human 3D genome architecture reveals chromatin topology for transcription. Cell, 2015, 163(7): 1611-1627.
doi: 10.1016/j.cell.2015.11.024 pmid: 26686651 |
| [14] | Osterwalder M, Barozzi I, Tissières V, Fukuda-Yuzawa Y, Mannion BJ, Afzal SY, Lee EA, Zhu YW, Plajzer-Frick I, Pickle CS, Kato M, Garvin TH, Pham QT, Harrington AN, Akiyama JA, Afzal V, Lopez-Rios J, Dickel DE, Visel A, Pennacchio LA. Enhancer redundancy provides phenotypic robustness in mammalian development. Nature, 2018, 554(7691): 239-243. |
| [15] |
Shlyueva D, Stelzer C, Gerlach D, Yáñez-Cuna JO, Rath M, Boryń ŁM, Arnold CD, Stark A. Hormone-responsive enhancer-activity maps reveal predictive motifs, indirect repression, and targeting of closed chromatin. Mol Cell, 2014, 54(1): 180-192.
doi: S1097-2765(14)00171-3 pmid: 24685159 |
| [16] | Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell, 1981, 27(2 Pt1): 299-308. |
| [17] |
Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell, 2013, 155(4): 934-947.
doi: 10.1016/j.cell.2013.09.053 pmid: 24119843 |
| [18] | Wu ZQ, Mi ZY. Research progress of super enhancer in cancer. Hereditas (Beijing), 2019, 41(1): 41-51. |
| 吴志强, 米泽云. 超级增强子在肿瘤研究中的进展. 遗传, 2019, 41(1): 41-51. | |
| [19] | Choukrallah MA, Song S, Rolink AG, Burger L, Matthias P. Enhancer repertoires are reshaped independently of early priming and heterochromatin dynamics during B cell differentiation. Nat Commun, 2015, 6: 8324. |
| [20] |
Samstein RM, Arvey A, Josefowicz SZ, Peng X, Reynolds A, Sandstrom R, Neph S, Sabo P, Kim JM, Liao W, Li MO, Leslie C, Stamatoyannopoulos JA, Rudensky AY. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell, 2012, 151(1): 153-166.
doi: 10.1016/j.cell.2012.06.053 pmid: 23021222 |
| [21] |
González AJ, Setty M, Leslie CS. Early enhancer establishment and regulatory locus complexity shape transcriptional programs in hematopoietic differentiation. Nat Genet, 2015, 47(11): 1249-1259.
doi: 10.1038/ng.3402 pmid: 26390058 |
| [22] | Xu CR, Cole PA, Meyers DJ, Kormish J, Dent S, Zaret KS. Chromatin "prepattern" and histone modifiers in a fate choice for liver and pancreas. Science, 2011, 332(6032): 963-966. |
| [23] | Visel A, Rubin EM, Pennacchio LA. Genomic views of distant-acting enhancers. Nature, 2009, 461(7261): 199-205. |
| [24] |
Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet, 2014, 15(4): 272-286.
doi: 10.1038/nrg3682 pmid: 24614317 |
| [25] |
Arnold PR, Wells AD, Li XC. Diversity and emerging roles of enhancer RNA in regulation of gene expression and cell fate. Front Cell Dev Biol, 2019, 7: 377.
doi: 10.3389/fcell.2019.00377 pmid: 31993419 |
| [26] |
Santiago-Algarra D, Dao LTM, Pradel L, España A, Spicuglia S. Recent advances in high-throughput approaches to dissect enhancer function. F1000Res, 2017, 6: 939.
doi: 10.12688/f1000research.11581.1 pmid: 28690838 |
| [27] | Liu Q, Li CY. The identification of enhancers and its application in cancer studies. Hereditas (Beijing), 2020, 42(9): 817-831. |
| 刘倩, 李春燕. 增强子的鉴定及其在肿瘤研究中的应用. 遗传, 2020, 42(9): 817-831. | |
| [28] | Ghavi-Helm Y, Klein FA, Pakozdi T, Ciglar L, Noordermeer D, Huber W, Furlong EEM. Enhancer loops appear stable during development and are associated with paused polymerase. Nature, 2014, 512(7512): 96-100. |
| [29] |
Benabdallah NS, Williamson I, Illingworth RS, Kane L, Boyle S, Sengupta D, Grimes GR, Therizols P, Bickmore WA. Decreased enhancer-promoter proximity accompanying enhancer activation. Mol Cell, 2019, 76(3): 473-484.e7.
doi: S1097-2765(19)30593-3 pmid: 31494034 |
| [30] | Alexander JM, Guan J, Li BK, Maliskova L, Song M, Shen Y, Huang B, Lomvardas S, Weiner OD. Live-cell imaging reveals enhancer-dependent Sox2 transcription in the absence of enhancer proximity. eLife, 2019, 8: e41769. |
| [31] | Thompson B, Varticovski L, Baek S, Hager GL. Genome-wide chromatin landscape transitions identify novel pathways in early commitment to osteoblast differentiation. PLoS One, 2016, 11(2): e0148619. |
| [32] | Luo LH, Gribskov M, Wang SF. Bibliometric review of ATAC-Seq and its application in gene expression. Brief Bioinform, 2022, 23(3): bbac061. |
| [33] | Zhou BR, Bai YW. Chromatin structures condensed by linker histones. Essays Biochem, 2019, 63(1): 75-87. |
| [34] |
Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng ZP, Furey TS, Crawford GE. High-resolution mapping and characterization of open chromatin across the genome. Cell, 2008, 132(2): 311-322.
doi: 10.1016/j.cell.2007.12.014 pmid: 18243105 |
| [35] | Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, Garg K, John S, Sandstrom R, Bates D, Boatman L, Canfield TK, Diegel M, Dunn D, Ebersol AK, Frum T, Giste E, Johnson AK, Johnson EM, Kutyavin T, Lajoie B, Lee BK, Lee K, London D, Lotakis D, Neph S, Neri F, Nguyen ED, Qu HZ, Reynolds AP, Roach V, Safi A, Sanchez ME, Sanyal A, Shafer A, Simon JM, Song LY, Vong S, Weaver M, Yan YQ, Zhang ZC, Zhang ZZ, Lenhard B, Tewari M, Dorschner MO, Hansen RS, Navas PA, Stamatoyannopoulos G, Iyer VR, Lieb JD, Sunyaev SR, Akey JM, Sabo PJ, Kaul R, Furey TS, Dekker J, Crawford GE, Stamatoyannopoulos JA. The accessible chromatin landscape of the human genome. Nature, 2012, 489(7414): 75-82. |
| [36] |
Zhao HN, Zhang WL, Zhang T, Lin Y, Hu YD, Fang C, Jiang JM. Genome-wide MNase hypersensitivity assay unveils distinct classes of open chromatin associated with H3K27me3 and DNA methylation in Arabidopsis thaliana. Genome Biol, 2020, 21(1): 24.
doi: 10.1186/s13059-020-1927-5 pmid: 32014062 |
| [37] |
He HH, Meyer CA, Hu SS, Chen MW, Zang CZ, Liu Y, Rao PK, Fei T, Xu H, Long H, Liu XS, Brown M. Refined DNase-seq protocol and data analysis reveals intrinsic bias in transcription factor footprint identification. Nat Methods, 2014, 11(1): 73-78.
doi: 10.1038/nmeth.2762 pmid: 24317252 |
| [38] |
Mckay DJ. Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to identify functional regulatory DNA in insect genomes. Methods Mol Biol, 2019, 1858: 89-97.
doi: 10.1007/978-1-4939-8775-7_8 pmid: 30414113 |
| [39] |
Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science, 2007, 316(5830): 1497-1502.
doi: 10.1126/science.1141319 pmid: 17540862 |
| [40] |
Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao YJ, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, Thiessen N, Griffith OL, He A, Marra M, Snyder M, Jones S. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods, 2007, 4(8): 651-657.
doi: 10.1038/nmeth1068 pmid: 17558387 |
| [41] |
Beacon TH, Delcuve GP, López C, Nardocci G, Kovalchuk I, van Wijnen AJ, Davie JR. The dynamic broad epigenetic (H3K4me3, H3K27ac) domain as a mark of essential genes. Clin Epigenetics, 2021, 13(1): 138.
doi: 10.1186/s13148-021-01126-1 pmid: 34238359 |
| [42] | Skene PJ, Henikoff S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife, 2017, 6: e21856. |
| [43] |
Kaya-Okur HS, Wu SJ, Codomo CA, Pledger ES, Bryson TD, Henikoff JG, Ahmad K, Henikoff S. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat Commun, 2019, 10(1): 1930.
doi: 10.1038/s41467-019-09982-5 pmid: 31036827 |
| [44] |
Kwasnieski JC, Fiore C, Chaudhari HG, Cohen BA. High-throughput functional testing of ENCODE segmentation predictions. Genome Res, 2014, 24(10): 1595-1602.
doi: 10.1101/gr.173518.114 pmid: 25035418 |
| [45] |
Lam MT, Li WB, Rosenfeld MG, Glass CK. Enhancer RNAs and regulated transcriptional programs. Trends Biochem Sci, 2014, 39(4): 170-182.
doi: 10.1016/j.tibs.2014.02.007 pmid: 24674738 |
| [46] |
Arnold CD, Gerlach D, Stelzer C, Boryń ŁM, Rath M, Stark A. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science, 2013, 339(6123): 1074-1077.
doi: 10.1126/science.1232542 pmid: 23328393 |
| [47] |
Muerdter F, Boryń ŁM, Arnold CD. STARR-seq— principles and applications. Genomics, 2015, 106(3): 145-150.
doi: S0888-7543(15)30010-0 pmid: 26072434 |
| [48] |
Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu Rev Biochem, 2003, 72: 449-479.
pmid: 12651739 |
| [49] |
Babbitt CC, Markstein M, Gray JM. Recent advances in functional assays of transcriptional enhancers. Genomics, 2015, 106(3): 137-139.
doi: S0888-7543(15)30005-7 pmid: 26100358 |
| [50] |
Muerdter F, Boryń ŁM, Woodfin AR, Neumayr C, Rath M, Zabidi MA, Pagani M, Haberle V, Kazmar T, Catarino RR, Schernhuber K, Arnold CD, Stark A. Resolving systematic errors in widely used enhancer activity assays in human cells. Nat Methods, 2018, 15(2): 141-149.
doi: 10.1038/nmeth.4534 pmid: 29256496 |
| [51] |
Peng TR, Zhai YN, Atlasi Y, Ter Huurne M, Marks H, Stunnenberg HG, Megchelenbrink W. STARR-seq identifies active, chromatin-masked, and dormant enhancers in pluripotent mouse embryonic stem cells. Genome Biol, 2020, 21(1): 243.
doi: 10.1186/s13059-020-02156-3 pmid: 32912294 |
| [52] | Wu YQ, Zhang YD, Liu H, Gao Y, Liu YY, Chen L, Liu L, Irwin DM, Hou CH, Zhou ZY, Zhang YP. Genome-wide identification of functional enhancers and their potential roles in pig breeding. J Anim Sci Biotechnol, 2022, 13(1): 75. |
| [53] |
Liu YW, Yu S, Dhiman VK, Brunetti T, Eckart H, White KP. Functional assessment of human enhancer activities using whole-genome STARR-sequencing. Genome Biol, 2017, 18(1): 219.
doi: 10.1186/s13059-017-1345-5 pmid: 29151363 |
| [54] | Sun JL, He N, Niu LJ, Huang YZ, Shen W, Zhang YD, Li L, Hou CH. Global quantitative mapping of enhancers in rice by STARR-seq. Genomics Proteomics Bioinformatics, 2019, 17(2): 140-153. |
| [55] |
Ntziachristos P, Abdel-Wahab O, Aifantis I. Emerging concepts of epigenetic dysregulation in hematological malignancies. Nat Immunol, 2016, 17(9): 1016-1024.
doi: 10.1038/ni.3517 pmid: 27478938 |
| [56] |
Liu F, Wang L, Perna F, Nimer SD. Beyond transcription factors: how oncogenic signalling reshapes the epigenetic landscape. Nat Rev Cancer, 2016, 16(6): 359-372.
doi: 10.1038/nrc.2016.41 pmid: 27220480 |
| [57] |
Gonda TJ, Ramsay RG. Directly targeting transcriptional dysregulation in cancer. Nat Rev Cancer, 2015, 15(11): 686-694.
doi: 10.1038/nrc4018 pmid: 26493648 |
| [58] |
Timp W, Feinberg AP. Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat Rev Cancer, 2013, 13(7): 497-510.
doi: 10.1038/nrc3486 pmid: 23760024 |
| [59] |
Zhang J, Lee D, Dhiman V, Jiang P, Xu J, Mcgillivray P, Yang HB, Liu J, Meyerson W, Clarke D, Gu MT, Li ST, Lou SK, Xu JR, Lochovsky L, Ung M, Ma LJ, Yu S, Cao Q, Harmanci A, Yan KK, Sethi A, Gürsoy G, Schoenberg MR, Rozowsky J, Warrell J, Emani P, Yang YT, Galeev T, Kong XM, Liu S, Li XT, Krishnan J, Feng YL, Rivera-Mulia JC, Adrian J, Broach JR, Bolt M, Moran J, Fitzgerald D, Dileep V, Liu TT, Mei SL, Sasaki T, Trevilla-Garcia C, Wang S, Wang YL, Zang CZ, Wang DF, Klein RJ, Snyder M, Gilbert DM, Yip K, Cheng C, Yue F, Liu XS, White KP, Gerstein M. An integrative ENCODE resource for cancer genomics. Nat Commun, 2020, 11(1): 3696.
doi: 10.1038/s41467-020-14743-w pmid: 32728046 |
| [60] |
Li QY, Seo JH, Stranger B, Mckenna A, Pe'er I, Laframboise T, Brown M, Tyekucheva S, Freedman ML. Integrative eQTL-based analyses reveal the biology of breast cancer risk loci. Cell, 2013, 152(3): 633-641.
doi: 10.1016/j.cell.2012.12.034 pmid: 23374354 |
| [61] |
Huang QL, Whitington T, Gao P, Lindberg JF, Yang YH, Sun JL, Väisänen MR, Szulkin R, Annala M, Yan J, Egevad LA, Zhang K, Lin RZ, Jolma A, Nykter M, Manninen A, Wiklund F, Vaarala MH, Visakorpi T, Xu JF, Taipale J, Wei GH. A prostate cancer susceptibility allele at 6q22 increases RFX6 expression by modulating HOXB13 chromatin binding. Nat Genet, 2014, 46(2): 126-135.
doi: 10.1038/ng.2862 pmid: 24390282 |
| [62] | Oldridge DA, Wood AC, Weichert-Leahey N, Crimmins I, Sussman R, Winter C, Mcdaniel LD, Diamond M, Hart LS, Zhu SZ, Durbin AD, Abraham BJ, Anders L, Tian LF, Zhang SL, Wei JS, Khan J, Bramlett K, Rahman N, Capasso M, Iolascon A, Gerhard DS, Guidry Auvil JM, Young RA, Hakonarson H, Diskin SJ, Look AT, Maris JM. Genetic predisposition to neuroblastoma mediated by a LMO1 super-enhancer polymorphism. Nature, 2015, 528(7582): 418-421. |
| [63] |
Zhang P, Xia JH, Zhu J, Gao P, Tian YJ, Du MJ, Guo YC, Suleman S, Zhang Q, Kohli M, Tillmans LS, Thibodeau SN, French AJ, Cerhan JR, Wang LD, Wei GH, Wang L. High-throughput screening of prostate cancer risk loci by single nucleotide polymorphisms sequencing. Nat Commun, 2018, 9(1): 2022.
doi: 10.1038/s41467-018-04451-x pmid: 29789573 |
| [64] |
Toropainen A, Stolze LK, Örd T, Whalen MB, Torrell PM, Link VM, Kaikkonen MU, Romanoski CE. Functional noncoding SNPs in human endothelial cells fine-map vascular trait associations. Genome Res, 2022, 32(3): 409-424.
doi: 10.1101/gr.276064.121 pmid: 35193936 |
| [65] | Chen XF, Duan YY, Jia YY, Dong QH, Shi W, Zhang Y, Dong SS, Li M, Liu ZB, Chen F, Huang XT, Hao RH, Zhu DL, Jing RH, Guo Y, Yang TL. Integrative high- throughput enhancer surveying and functional verification divulges a YY2-condensed regulatory axis conferring risk for osteoporosis. Cell Genom, 2024, 4(3): 100501. |
| [66] | Hansen TJ, Hodges E. ATAC-STARR-seq reveals transcription factor-bound activators and silencers across the chromatin accessible human genome. Genome Res, 2022, 32(8): 1529-1541. |
| [67] |
Glaser LV, Steiger M, Fuchs A, van Bömmel A, Einfeldt E, Chung HR, Vingron M, Meijsing SH. Assessing genome-wide dynamic changes in enhancer activity during early mESC differentiation by FAIRE-STARR-seq. Nucleic Acids Res, 2021, 49(21): 12178-12195.
doi: 10.1093/nar/gkab1100 pmid: 34850108 |
| [68] | Chan YC, Kienle E, Oti M, Di Liddo A, Mendez-Lago M, Aschauer DF, Peter M, Pagani M, Arnold C, Vonderheit A, Schön C, Kreuz S, Stark A, Rumpel S. An unbiased AAV-STARR-seq screen revealing the enhancer activity map of genomic regions in the mouse brain in vivo. Sci Rep, 2023, 13(1): 6745. |
| [69] |
Barakat TS, Halbritter F, Zhang M, Rendeiro AF, Perenthaler E, Bock C, Chambers I. Functional dissection of the enhancer repertoire in human embryonic stem cells. Cell Stem Cell, 2018, 23(2): 276-288.e8.
doi: S1934-5909(18)30296-0 pmid: 30033119 |
| [70] |
Sidik SM, Huet D, Lourido S. CRISPR-Cas9-based genome-wide screening of Toxoplasma gondii. Nat Protoc, 2018, 13(1): 307-323.
doi: 10.1038/nprot.2017.131 pmid: 29323662 |
| [71] | Zhao YM, Wang F, Wang CH, Zhang XB, Jiang CZ, Ding F, Shen L, Zhang QF. Optimization of CRISPR/Cas system for improving genome editing efficiency in plasmodium falciparum. Front Microbiol, 2020, 11: 625862. |
| [72] | Zhang WW, Lypaczewski P, Matlashewski G. Application of CRISPR/Cas9-mediated genome editing in Leishmania. Methods Mol Biol, 2020, 2116: 199-224. |
| [73] |
Bachmaier S, Thanner T, Boshart M. Culturing and transfection of pleomorphic Trypanosoma brucei. Methods Mol Biol, 2020, 2116: 23-38.
doi: 10.1007/978-1-0716-0294-2_2 pmid: 32221911 |
| [74] | Chen L, Fish AE, Capra JA. Prediction of gene regulatory enhancers across species reveals evolutionarily conserved sequence properties. PLoS Comput Biol, 2018, 14(10): e1006484. |
| [75] |
Chaudhri VK, Dienger-Stambaugh K, Wu ZG, Shrestha M, Singh H. Charting the cis-regulome of activated B cells by coupling structural and functional genomics. Nat Immunol, 2020, 21(2): 210-220.
doi: 10.1038/s41590-019-0565-0 pmid: 31873292 |
| [1] | 安梦婷, 郭冠麟, 吴杰, 孙文靖, 贾学渊. 基于生物信息学分析胃癌双微体中增强子的调控机制[J]. 遗传, 2025, 47(5): 558-572. |
| [2] | 李轲, 周晓蓉, 朱东丽, 陈晓峰, 郭燕. 系统性红斑狼疮易感区域FAM167A-BLK遗传变异的调控机制研究[J]. 遗传, 2025, 47(11): 1244-1255. |
| [3] | 任佳琳, 童依泽, 蔡瑞. 染色质相关RNA的m6A修饰调控染色质可及性与基因转录研究进展[J]. 遗传, 2025, 47(11): 1186-1196. |
| [4] | 杨敏, 林思远, 杨长淇, 陈瑶生, 何祖勇. SOX9及其增强子在哺乳动物性别决定中的研究进展[J]. 遗传, 2024, 46(9): 677-689. |
| [5] | 尤琳琳, 张余. 细菌转录终止的分子机制研究进展[J]. 遗传, 2024, 46(12): 982-994. |
| [6] | 王承贤, 容益康, 崔敏. 果蝇限制端粒转座子的分子机制[J]. 遗传, 2023, 45(3): 221-228. |
| [7] | 陈秀丽, 黄海燕, 吴强. 靶向敲除β-珠蛋白基因座控制区增强子HS2对K562细胞转录组的影响[J]. 遗传, 2022, 44(9): 783-797. |
| [8] | 吴丹丹, 朱明昆, 方忠艳, 马伟. 植物B染色体的分子结构组成及遗传机制研究进展[J]. 遗传, 2022, 44(9): 772-782. |
| [9] | 徐思远, 寿佳, 吴强. HS5-1增强子eRNA PEARL对原钙粘蛋白α基因簇的表达调控[J]. 遗传, 2022, 44(8): 695-764. |
| [10] | 张元, 赵语婷, 庄乐南, 贺津. 转录中介体复合物在心血管发育和疾病中的转录调控作用[J]. 遗传, 2022, 44(5): 383-397. |
| [11] | 漆思晗, 王棨临, 张俊有, 刘倩, 李春燕. 增强子调控癌症发生发展的机制研究[J]. 遗传, 2022, 44(4): 275-288. |
| [12] | 万星琦, 魏婉珍, 郭胜良, 崔一笑, 景雪莹, 黄露杰, 马捷. BMP2基因远程调控元件的功能分析[J]. 遗传, 2022, 44(12): 1141-1147. |
| [13] | 刘国芳, 任沛东, 叶文新, 陆光涛. 十字花科黑腐病菌中转录因子HpaR1与Clp调控一个糖苷水解酶基因表达的分析[J]. 遗传, 2021, 43(9): 910-920. |
| [14] | 王玲, 李金环, 黄海燕, 吴强. 串联反向CTCF位点的系列删除揭示增强子调控HOXD基因簇表达的平衡[J]. 遗传, 2021, 43(8): 775-791. |
| [15] | 王天一, 王应祥, 尤辰江. 植物PHD结构域蛋白的结构与功能特性[J]. 遗传, 2021, 43(4): 323-339. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||

www.chinagene.cn
备案号:京ICP备09063187号-4
总访问:,今日访问:,当前在线:
