遗传 ›› 2020, Vol. 42 ›› Issue (10): 929-948.doi: 10.16288/j.yczz.20-107
收稿日期:2020-04-18
修回日期:2020-05-06
出版日期:2020-10-20
发布日期:2020-05-25
通讯作者:
王龙
E-mail:171240503@smail.nju.edu.cn;wanglong@nju.edu.cn
作者简介:高志伟,在读本科生,专业方向:生物化学与分子生物学。E-mail: 基金资助:Received:2020-04-18
Revised:2020-05-06
Online:2020-10-20
Published:2020-05-25
Contact:
Wang Long
E-mail:171240503@smail.nju.edu.cn;wanglong@nju.edu.cn
Supported by:摘要:
作为重大进化谜题,真核生物起源的研究对于解码真核基因组、阐释真核细胞内部结构之间的关系有重要启示作用。在1977年美国微生物学家Carl Woese发现古细菌并提出三域生命之树之后,大量研究显示古细菌与真核生物在进化上存在着密切联系。21世纪以来,系统发育分析方法不断改进,泉古菌门(Crenarchaeota)、广古菌门(Euryarchaeota)之外与真核生物更加相似的新古细菌门类也相继被发现,这些证据更加支持将真核生物与古细菌合并为一域,形成二域生命之树。目前,通过宏基因组技术发现的Asgard古细菌是与真核生物进化距离最近的原核生物。然而,真核生物祖先的身份以及线粒体起源的时间等核心问题仍是学术界争论的焦点。本文结合近年来国内外研究成果,从生命之树的形态变化与真核生物演变的具体机制两个角度梳理了目前对真核生物起源的认知过程、现有水平和研究前景,以期为揭示真核生物起源进程的后续研究提供参考与指引。
高志伟, 王龙. 真核生物起源研究进展[J]. 遗传, 2020, 42(10): 929-948.
Zhiwei Gao, Long Wang. Progress in elucidating the origin of eukaryotes[J]. Hereditas(Beijing), 2020, 42(10): 929-948.
表1
2003~2009年间发表的7篇阐释生命之树中古细菌与真核生物关系的文章比较"
| 文献 | 三域中使用的标记数目 | 取样种类(数目) | 氨基酸位置数 | 方法 | 结果 | 优势 | 缺陷 |
|---|---|---|---|---|---|---|---|
| Harris等[ | 50 | 细菌(25); 泉古菌门(1); 广古菌门(7); 真核生物(3) | 与所分析基因相关 | 单基因分析; 最大似然法; 最大简约法; 距离法 | 3D | 选取数据范围从此前的SSU rRNA扩展到其 他基因 | 当时的基因数据有限,单基因分析可靠性不强 |
| Ciccarelli 等[ | 31 | 细菌(150); 泉古菌门(4); 广古菌门(14); 真核生物(23) | 8090 | 分散基因串联 | 3D | 取样物种广泛, 增加了客观的 HGT过滤方法 | 先在域内对齐序列,跨域整合时可能将非同源基因对齐 |
| Yutin等[ | 136 | 与所用基因 相关 | 与所分析基因相关 | 单基因分析; 最大似然法 | 3D | 数据集丰富,分析的基因数目多 | 单个基因提供的系统发育信号弱 |
| Rivera和Lake[ | Archaeoglobus fulgidus全基因组 | 细菌(2); 泉古菌门(1); 广古菌门(2); 真核生物(2) | 不适用 | 基因组条件 重建 | 2D; 真核生物与泉古菌门为姐妹群 | 使用全基因组 分析 | 取样物种少,受HGT和基因丢失影响大 |
| Pisani等[ | 未提供数据 | 细菌(97); 泉古菌门(4); 广古菌门(17); 真核生物(17) | 与所分析基因相关 | 超树法 (Supertree) | 2D; 真核生物与广古菌门为姐妹群 | 综合众多数据集信息,在单基因 树基础上建立二级树 | 过滤步骤后剩余的有效数据少,且拥有单基因分析的局限性 |
| Cox等[ | 45 | 细菌(10); 泉古菌门(3); 广古菌门(11); 真核生物(16) | 5521 | 分散基因串联; 贝叶斯方法; 最大似然法; 最大简约法 | 2D; 真核生物与泉古菌门为姐妹群 | 除简单模型外使用了更多更复杂的模型,首例使用新模型证明了2D系统结果 | 复杂模型的参数估计的精确性难以保证 |
| Foster等[ | 41 | 细菌(8); 泉古菌门(8); 奇古菌门(2); 广古菌门(6); 真核生物(11) | 5222 | 分散基因串联; 贝叶斯方法; 最大似然法; 最大简约法 | 2D; 真核生物是一个由泉古菌门和奇古菌门构成的群的姐妹群 | 同上,作者相同,选取的数据更多,使用了新发现的古细菌门类数据 | 复杂模型的参数估计的精确性难以保证 |
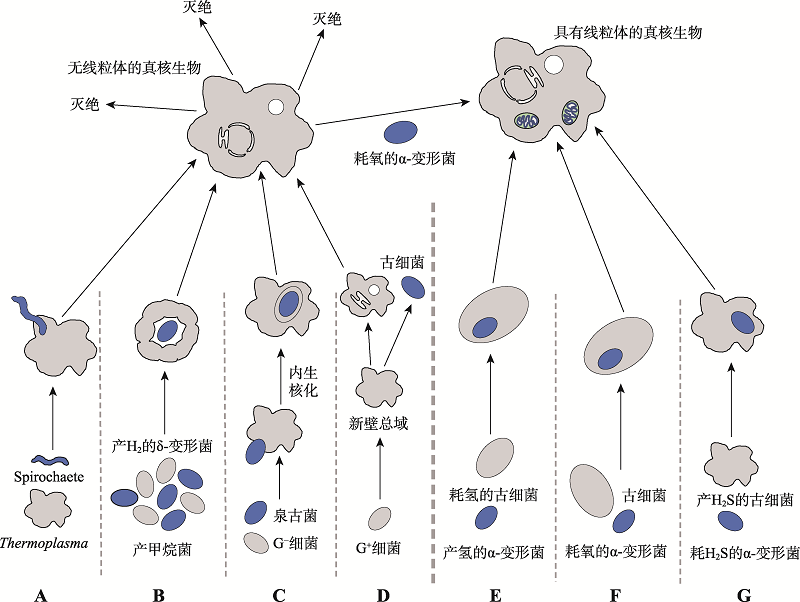
图7
有关线粒体起源的模型 A~D为晚期线粒体模型,E~G为早期线粒体模型。A:Margulis认为古细菌宿主热原体菌(Thermoplasma)首先与螺旋菌(Spirochaete)内共生,逐渐演化出细胞核,后形成线粒体[100]。B:互养共栖假说,产甲烷古细菌与δ-变形菌在代谢上存在氢的交换。一个产甲烷菌被多个δ-变形菌包围后,δ-变形菌间的相互作用促进胞质融合,形成内质网、细胞核等结构,δ-变形菌基因组随着基因的转移逐渐消失,线粒体的内共生最后发生[32]。C:内生核假说,泉古菌与G-真细菌互相作用,泉古菌形成细胞核,真细菌形成细胞质,再与α-变形菌内共生形成线粒体[101]。D:Cavalier-Smith认为真核生物与古细菌演化自一个共同祖先,同属新壁总域[97,99]。E:氢假说,宿主是严格依赖氢且严格自养的厌氧古细菌,共生体能进行有氧呼吸,但通过厌氧异养代谢为宿主提供氢气[31]。F:古细菌宿主在形成真核细胞的其他复杂结构前首先与耗氧的α-变形菌内共生形成线粒体[103]。G:基于硫元素的代谢循环,宿主是产H2S的古细菌,线粒体祖先是耗H2S的α-变形菌[104]。根据文献[30]修改绘制。"
| [1] | Blackstone NW . An evolutionary framework for understanding the origin of eukaryotes. Biology (Basel), 2016,5(2):18. |
| [2] |
Gabaldón T . Relative timing of mitochondrial endosymbiosis and the “pre-mitochondrial symbioses” hypothesis. IUBMB Life, 2018,70(12):1188-1196.
doi: 10.1002/iub.1950 pmid: 30358047 |
| [3] |
Fuerst JA . Intracellular compartmentation in planctomycetes. Annu Rev Microbiol, 2005,59:299-328.
doi: 10.1146/annurev.micro.59.030804.121258 pmid: 15910279 |
| [4] |
Martin W, Koonin EV . Introns and the origin of nucleus-cytosol compartmentalization. Nature, 2006,440(7080):41-45.
doi: 10.1038/nature04531 pmid: 16511485 |
| [5] | Lin L, Shi AB . Endocytic recycling pathways and the regulatory mechanisms. Hereditas(Beijing), 2019,41(6):451-468. |
| 林珑, 史岸冰 . 细胞内吞循环运输通路及其分子调控机制. 遗传, 2019,41(6):451-468. | |
| [6] |
Dacks JB, Field MC, Buick R, Eme L, Gribaldo S, Roger AJ, Brochier-Armanet C, Devos DP . The changing view of eukaryogenesis-fossils, cells, lineages and how they all come together. J Cell Sci, 2016,129(20):3695-3703.
doi: 10.1242/jcs.178566 pmid: 27672020 |
| [7] |
Shen Y, Buick R, Canfield DE . Isotopic evidence for microbial sulphate reduction in the early Archaean era. Nature, 2001,410(6824):77-81.
doi: 10.1038/35065071 pmid: 11242044 |
| [8] |
Ueno Y, Ono S, Rumble D, Maruyama S . Quadruple sulfur isotope analysis of ca. 3.5 Ga Dresser Formation: New evidence for microbial sulfate reduction in the early Archean. Geochim Cosmochim Ac, 2008,72(23):5675-5691.
doi: 10.1016/j.gca.2008.08.026 |
| [9] | Yan YZ, Liu ZL . Significance of eucaryotic organisms in the microfossil flora of Changcheng system. Acta Micropalaeontol Sin, 1993,10(2):167-180. |
| [10] |
Parfrey LW, Lahr DJG, Knoll AH, Katz LA . Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc Natl Acad Sci USA, 2011,108(33):13624-13629.
doi: 10.1073/pnas.1110633108 pmid: 21810989 |
| [11] |
Spang A, Saw JH, Jørgensen SL, Zaremba-Niedzwiedzka K, Martijn J, Lind AE, van Eijk R, Schleper C, Guy L, Ettema TJG,. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature, 2015,521(7551):173-179.
pmid: 25945739 |
| [12] |
Zaremba-Niedzwiedzka K, Caceres EF, Saw JH, Bäckström D, Juzokaite L, Vancaester E, Seitz KW, Anantharaman K, Starnawski P, Kjeldsen KU, Stott MB, Nunoura T, Banfield JF, Schramm A, Baker BJ, Spang A, Ettema TJ . Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature, 2017,541(7637):353-358.
doi: 10.1038/nature21031 pmid: 28077874 |
| [13] |
Imachi H, Nobu MK, Nakahara N, Morono Y, Ogawara M, Takaki Y, Takano Y, Uematsu K, Ikuta T, Ito M, Matsui Y, Miyazaki M, Murata K, Saito K, Sakai S, Song CH, Tasumi E, Yamanaka Y, Yamaguchi T, Kamagata Y, Tamaki H, Takai K . Isolation of an archaeon at the prokaryote-eukaryote interface. Nature, 2020,577(7791):519-525.
doi: 10.1038/s41586-019-1916-6 pmid: 31942073 |
| [14] |
Woese CR, Fox GE . Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci USA, 1977,74(11):5088-5090.
pmid: 270744 |
| [15] |
Iwabe N, Kuma K, Hasegawa M, Osawa S, Miyata T . Evolutionary relationship of archaebacteria, eubacteria, and eukaryotes inferred from phylogenetic trees of duplicated genes. Proc Natl Acad Sci USA, 1989,86(23):9355-9359.
doi: 10.1073/pnas.86.23.9355 |
| [16] |
Woese CR, Kandler O, Wheelis ML . Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA, 1990,87(12):4576-4579.
doi: 10.1073/pnas.87.12.4576 pmid: 2112744 |
| [17] |
Eme L, Spang A, Lombard J, Stairs CW, Ettema TJG . Archaea and the origin of eukaryotes. Nat Rev Microbiol, 2017,15(12):711-723.
doi: 10.1038/nrmicro.2017.133 pmid: 29123225 |
| [18] |
Pace NR . Time for a change. Nature, 2006,441(7091):289-289.
doi: 10.1038/441289a pmid: 16710401 |
| [19] |
Huet J, Schnabel R, Sentenac A, Zillig W . Archaebacteria and eukaryotes possess DNA‐dependent RNA polymerases of a common type. EMBO J, 1983,2(8):1291-1294.
pmid: 10872322 |
| [20] |
Zillig W, Klenk HP, Palm P, Pühler G, Gropp F, Garrett RA, Leffers H . The phylogenetic relations of DNA- dependent RNA polymerases of archaebacteria, eukaryotes, and eubacteria. Can J Microbiol, 1989,35(1):73-80.
doi: 10.1139/m89-011 pmid: 2541879 |
| [21] |
Gribaldo S, Poole AM, Daubin V, Forterre P, Brochier-Armanet C . The origin of eukaryotes and their relationship with the Archaea: are we at a phylogenomic impasse? Nat Rev Microbiol, 2010,8(10):743-752.
pmid: 20844558 |
| [22] |
Lindås AC, Karlsson EA, Lindgren MT, Ettema TJ, Bernander R . A unique cell division machinery in the Archaea. Proc Natl Acad Sci USA, 2008,105(48):18942-18946.
doi: 10.1073/pnas.0809467105 pmid: 18987308 |
| [23] |
Samson RY, Obita T, Freund SM, Williams RL, Bell SD . A role for the ESCRT system in cell division in archaea. Science, 2008,322(5908):1710-1713.
doi: 10.1126/science.1165322 pmid: 19008417 |
| [24] | Li J, Zhang KQ . Genetic diversity of microorganisms. Hereditas(Beijing), 2012,34(11):1399-1408. |
| 李娟, 张克勤 . 微生物的遗传多样性. 遗传, 2012,34(11):1399-1408. | |
| [25] |
Rivera MC, Jain R, Moore JE, Lake JA . Genomic evidence for two functionally distinct gene classes. Proc Natl Acad Sci USA, 1998,95(11):6239-6244.
doi: 10.1073/pnas.95.11.6239 pmid: 9600949 |
| [26] |
Canback B, Andersson SG, Kurland CG . The global phylogeny of glycolytic enzymes. Proc Natl Acad Sci USA, 2002,99(9):6097-6102.
doi: 10.1073/pnas.082112499 pmid: 11983902 |
| [27] |
Peretó J, López-García P, Moreira D . Ancestral lipid biosynthesis and early membrane evolution. Trends Biochem Sci, 2004,29(9):469-477.
doi: 10.1016/j.tibs.2004.07.002 pmid: 15337120 |
| [28] |
Lake JA, Henderson E, Oakes M, Clark MW . Eocytes: a new ribosome structure indicates a kingdom with a close relationship to eukaryotes. Proc Natl Acad Sci USA, 1984,81(12):3786-3790.
doi: 10.1073/pnas.81.12.3786 pmid: 6587394 |
| [29] |
Rivera MC, Lake JA . Evidence that eukaryotes and eocyte prokaryotes are immediate relatives. Science, 1992,257(5066):74-76.
pmid: 1621096 |
| [30] |
Embley TM, Martin W . Eukaryotic evolution, changes and challenges. Nature, 2006,440(7084):623-630.
doi: 10.1038/nature04546 pmid: 16572163 |
| [31] |
Martin W, Müller M . The hydrogen hypothesis for the first eukaryote. Nature, 1998,392(6671):37-41.
doi: 10.1038/32096 pmid: 9510246 |
| [32] |
Moreira D, López-García P . Symbiosis between methanogenic archaea and δ-proteobacteria as the origin of eukaryotes: the syntrophic hypothesis. J Mol Evol, 1998,47(5):517-530.
doi: 10.1007/pl00006408 pmid: 9797402 |
| [33] |
Harris JK, Kelley ST, Spiegelman GB, Pace NR . The genetic core of the universal ancestor. Genome Res, 2003,13(3):407-412.
doi: 10.1101/gr.652803 pmid: 12618371 |
| [34] |
Ciccarelli FD, Doerks T, Von Mering C, Creevey CJ, Snel B, Bork P . Toward automatic reconstruction of a highly resolved tree of life. Science, 2006,311(5765):1283-1287.
doi: 10.1126/science.1123061 pmid: 16513982 |
| [35] |
Yutin N, Makarova KS, Mekhedov SL, Wolf YI, Koonin EV . The deep archaeal roots of eukaryotes. Mol Biol Evol, 2008,25(8):1619-1630.
pmid: 18463089 |
| [36] |
Rivera MC, Lake JA . The ring of life provides evidence for a genome fusion origin of eukaryotes. Nature, 2004,431(7005):152-155.
doi: 10.1038/nature02848 pmid: 15356622 |
| [37] |
Pisani D, Cotton JA , McInerney JO. Supertrees disentangle the chimerical origin of eukaryotic genomes. Mol Biol Evol, 2007,24(8):1752-1760.
pmid: 17504772 |
| [38] |
Cox CJ, Foster PG, Hirt RP, Harris SR, Embley TM . The archaebacterial origin of eukaryotes. Proc Natl Acad Sci USA, 2008,105(51):20356-20361.
doi: 10.1073/pnas.0810647105 pmid: 19073919 |
| [39] |
Foster PG, Cox CJ, Embley TM . The primary divisions of life: a phylogenomic approach employing composition- heterogeneous methods. Philos Trans R Soc Lond B Biol Sci, 2009,364(1527):2197-2207.
doi: 10.1098/rstb.2009.0034 pmid: 19571240 |
| [40] |
Boussau B, Daubin V . Genomes as documents of evolutionary history. Trends Ecol Evol, 2010,25(4):224-232.
doi: 10.1016/j.tree.2009.09.007 |
| [41] |
Som A . Causes, consequences and solutions of phylogenetic incongruence. Brief Bioinform, 2015,16(3):536-548.
doi: 10.1093/bib/bbu015 pmid: 24872401 |
| [42] |
Lerat E, Daubin V, Ochman H, Moran NA . Evolutionary origins of genomic repertoires in bacteria. PLoS Biol, 2005,3(5):e130.
doi: 10.1371/journal.pbio.0030130 pmid: 15799709 |
| [43] |
Soucy SM, Huang J, Gogarten JP . Horizontal gene transfer: building the web of life. Nat Rev Genet, 2015,16(8):472-482.
doi: 10.1038/nrg3962 pmid: 26184597 |
| [44] |
Wagner A, Whitaker RJ, Krause DJ, Heilers JH, Van Wolferen M, Van Der Does C, Albers SV,. Mechanisms of gene flow in archaea. Nat Rev Microbiol, 2017,15(8):492-501.
doi: 10.1038/nrmicro.2017.41 pmid: 28502981 |
| [45] |
Akanni WA, Siu-Ting K, Creevey CJ, McInerney JO, Wilkinson M, Foster PG, Pisani D,. Horizontal gene flow from Eubacteria to Archaebacteria and what it means for our understanding of eukaryogenesis. Philos Trans R Soc Lond B Biol Sci, 2015,370(1678):20140337.
doi: 10.1098/rstb.2014.0337 pmid: 26323767 |
| [46] |
Cotton JA, McInerney JO. Eukaryotic genes of archaebacterial origin are more important than the more numerous eubacterial genes, irrespective of function. Proc Natl Acad Sci USA, 2010,107(40):17252-17255.
doi: 10.1073/pnas.1000265107 pmid: 20852068 |
| [47] |
Cohen O, Gophna U, Pupko T . The complexity hypothesis revisited: connectivity rather than function constitutes a barrier to horizontal gene transfer. Mol Biol Evol, 2011,28(4):1481-1489.
doi: 10.1093/molbev/msq333 |
| [48] |
Williams TA, Foster PG, Cox CJ, Embley TM . An archaeal origin of eukaryotes supports only two primary domains of life. Nature, 2013,504(7479):231-236.
doi: 10.1038/nature12779 |
| [49] |
Williams TA, Cox CJ, Foster PG, Szöllősi GJ, Embley TM . Phylogenomics provides robust support for a two-domains tree of life. Nat Ecol Evol, 2020,4(1):138-147.
doi: 10.1038/s41559-019-1040-x pmid: 31819234 |
| [50] |
Tourasse NJ, Gouy M . Accounting for evolutionary rate variation among sequence sites consistently changes universal phylogenies deduced from rRNA and protein-coding genes. Mol Phylogenet Evol, 1999,13(1):159-168.
doi: 10.1006/mpev.1999.0675 pmid: 10508549 |
| [51] |
Guy L, Ettema TJ . The archaeal ‘TACK’ superphylum and the origin of eukaryotes. Trends Microbiol, 2011,19(12):580-587.
doi: 10.1016/j.tim.2011.09.002 |
| [52] |
Williams TA, Foster PG, Nye TM, Cox CJ, Embley TM . A congruent phylogenomic signal places eukaryotes within the Archaea. Proc Biol Sci, 2012,279(1749):4870-4879.
doi: 10.1098/rspb.2012.1795 pmid: 23097517 |
| [53] |
Rochette NC, Brochier-Armanet C, Gouy M . Phylogenomic test of the hypotheses for the evolutionary origin of eukaryotes. Mol Biol Evol, 2014,31(4):832-845.
doi: 10.1093/molbev/mst272 |
| [54] |
Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers YH, Smith HO . Environmental genome shotgun sequencing of the Sargasso Sea. Science, 2004,304(5667):66-74.
doi: 10.1126/science.1093857 pmid: 15001713 |
| [55] |
Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P . Mesophilic Crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol, 2008,6(3):245-252.
doi: 10.1038/nrmicro1852 pmid: 18274537 |
| [56] |
Elkins JG, Podar M, Graham DE, Makarova KS, Wolf Y, Randau L, Hedlund BP, Brochier-Armanet C, Kunin V, Anderson I, apidus A, Goltsman E, Barry K, Koonin EV, Hugenholtz P, Kyrpides N, Wanner G, Richardson P, Keller M, Stettera K. A korarchaeal genome reveals insights into the evolution of the Archaea. Proc Natl Acad Sci USA, 2008,105(23):8102-8107.
doi: 10.1073/pnas.0801980105 pmid: 18535141 |
| [57] |
Nunoura T, Takaki Y, Kakuta J, Nishi S, Sugahara J, Kazama H, Chee GJ, Hattori M, Kanai A, Atomi H, Takai K, Takami H . Insights into the evolution of Archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic Acids Res, 2011,39(8):3204-3223.
doi: 10.1093/nar/gkq1228 pmid: 21169198 |
| [58] |
Guy L, Saw JH, Ettema TJ . The archaeal legacy of eukaryotes: a phylogenomic perspective. Cold Spring Harb Perspect Biol, 2014,6(10):a016022.
doi: 10.1101/cshperspect.a016022 pmid: 24993577 |
| [59] |
Petitjean C, Deschamps P, López-García P, Moreira D . Rooting the domain archaea by phylogenomic analysis supports the foundation of the new kingdom Proteoarchaeota. Genome Biol Evol, 2015,7(1):191-204.
doi: 10.1093/gbe/evu274 |
| [60] |
Raymann K, Brochier-Armanet C, Gribaldo S . The two-domain tree of life is linked to a new root for the Archaea. Proc Natl Acad Sci USA, 2015,112(21):6670-6675.
doi: 10.1073/pnas.1420858112 pmid: 25964353 |
| [61] |
MacLeod F, Kindler GS, Wong HL, Chen R, Burns BP . Asgard archaea: diversity, function, and evolutionary implications in a range of microbiomes. AIMS Microbiol, 2019,5(1):48-61.
doi: 10.3934/microbiol.2019.1.48 pmid: 31384702 |
| [62] |
Seitz KW, Lazar CS, Hinrichs KU, Teske AP, Baker BJ . Genomic reconstruction of a novel, deeply branched sediment archaeal phylum with pathways for acetogenesis and sulfur reduction. ISME J, 2016,10(7):1696-1705.
doi: 10.1038/ismej.2015.233 pmid: 26824177 |
| [63] |
López-García P, Moreira D . Eukaryogenesis, a syntrophy affair. Nat Microbiol, 2019,4(7):1068-1070.
doi: 10.1038/s41564-019-0495-5 pmid: 31222170 |
| [64] |
Da Cunha V, Gaia M, Gadelle D, Nasir A, Forterre P . Lokiarchaea are close relatives of Euryarchaeota, not bridging the gap between prokaryotes and eukaryotes. PLoS Genet, 2017,13(6):e1006810.
doi: 10.1371/journal.pgen.1006810 pmid: 28604769 |
| [65] |
Spang A, Eme L, Saw JH, Caceres EF, Zaremba- Niedzwiedzka K, Lombard J, Guy L, Ettema TJG . Asgard archaea are the closest prokaryotic relatives of eukaryotes. PLoS Genet, 2018,14(3):e1007080.
doi: 10.1371/journal.pgen.1007080 |
| [66] |
Da Cunha V, Gaia M, Nasir A, Forterre P . Asgard archaea do not close the debate about the universal tree of life topology. PLoS Genet, 2018,14(3):e1007215.
doi: 10.1371/journal.pgen.1007215 pmid: 29596428 |
| [67] |
Narrowe AB, Spang A, Stairs CW, Caceres EF, Baker BJ, Miller CS, Ettema TJG . Complex evolutionary history of translation Elongation Factor 2 and diphthamide biosynthesis in Archaea and parabasalids. Genome Biol Evol, 2018,10(9):2380-2393.
doi: 10.1093/gbe/evy154 pmid: 30060184 |
| [68] |
Harish A, Tunlid A, Kurland CG . Rooted phylogeny of the three superkingdoms. Biochimie, 2013,95(8):1593-1604.
doi: 10.1016/j.biochi.2013.04.016 |
| [69] |
Harish A, Kurland CG . Akaryotes and Eukaryotes are independent descendants of a universal common ancestor. Biochimie, 2017,138:168-183.
doi: 10.1016/j.biochi.2017.04.013 pmid: 28461155 |
| [70] |
Harish A . What is an archaeon and are the Archaea really unique? PeerJ, 2018,6:e5770.
doi: 10.7717/peerj.5770 pmid: 30357005 |
| [71] |
Caetano-Anollés G, Caetano-Anollés D . An evolutionarily structured universe of protein architecture. Genome Res, 2003,13(7):1563-1571.
pmid: 12840035 |
| [72] |
Yang S, Doolittle RF, Bourne PE . Phylogeny determined by protein domain content. Proc Natl Acad Sci USA, 2005,102(2):373-378.
doi: 10.1073/pnas.0408810102 pmid: 15630082 |
| [73] |
Harish A, Morrison DA . The deep(er) roots of Eukaryotes and Akaryotes. F1000 Res, 2020,9(112):112.
doi: 10.12688/f1000research |
| [74] |
Fournier GP, Poole AM . A briefly argued case that Asgard archaea are part of the eukaryote tree. Front Microbiol, 2018,9:1896.
doi: 10.3389/fmicb.2018.01896 pmid: 30158917 |
| [75] |
Seitz KW, Dombrowski N, Eme L, Spang A, Lombard J, Sieber JR, Teske AP, Ettema TJ, Baker BJ . Asgard archaea capable of anaerobic hydrocarbon cycling. Nat Commun, 2019,10(1):1-11.
pmid: 30602773 |
| [76] | Cai MW, Liu Y, Zhou ZC, Yang YC, Pan J, Gu JD, Li M . Asgard archaea are diverse, ubiquitous, and transcriptionally active microbes. bioRxiv, 2018: 374165. |
| [77] | Cai MW, Liu Y, Yin XR, Zhou ZC, Friedrich MW, Richter-Heitmann T, Nimzyk R, Kulkarni A, Wang XW, Li WJ, Pan J, Yang YC, Gu JD, Li M . Highly diverse Asgard archaea participate in organic matter degradation in coastal sediments. bioRxiv, 2019: 858530. |
| [78] |
Zhang RY, Zou B, Yan YW, Jeon CO, Li M, Cai M, Quan ZX . Design of targeted primers based on 16S rRNA sequences in meta-transcriptomic datasets and identification of a novel taxonomic group in the Asgard archaea. BMC Microbiol, 2020,20(1):25.
doi: 10.1186/s12866-020-1707-0 pmid: 32013868 |
| [79] | Caceres EF . Genomic and evolutionary exploration of Asgard archaea. Acta Univ Ups, 2019, 60-61. |
| [80] |
Koonin EV . The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol, 2010,11(5):209.
doi: 10.1186/gb-2010-11-5-209 pmid: 20441612 |
| [81] |
Koumandou VL, Wickstead B, Ginger ML, Van Der Giezen M, Dacks JB, Field MC. Molecular paleontology and complexity in the last eukaryotic common ancestor. Crit Rev Biochem Mol Biol, 2013,48(4):373-396.
doi: 10.3109/10409238.2013.821444 pmid: 23895660 |
| [82] |
Rout MP, Field MC . The evolution of organellar coat complexes and organization of the eukaryotic cell. Annu Rev Biochem, 2017,86:637-657.
doi: 10.1146/annurev-biochem-061516-044643 |
| [83] |
Gould SB . Membranes and evolution. Curr Biol, 2018,28(8):R381-R385.
doi: 10.1016/j.cub.2018.01.086 pmid: 29689219 |
| [84] |
López-García P, Moreira D . Open questions on the origin of eukaryotes. Trends Ecol Evol, 2015,30(11):697-708.
doi: 10.1016/j.tree.2015.09.005 pmid: 26455774 |
| [85] |
Gray MW . Mosaic nature of the mitochondrial proteome: Implications for the origin and evolution of mitochondria. Proc Natl Acad Sci USA, 2015,112(33):10133-10138.
doi: 10.1073/pnas.1421379112 pmid: 25848019 |
| [86] |
Santos HJ, Makiuchi T, Nozaki T . Reinventing an organelle: the reduced mitochondrion in parasitic protists. Trends Parasitol, 2018,34(12):1038-1055.
doi: 10.1016/j.pt.2018.08.008 pmid: 30201278 |
| [87] | Tachezy J . Hydrogenosomes and mitosomes: mitochondria of anaerobic eukaryotes. Springer, 2019, 1-20. |
| [88] |
Karnkowska A, Vacek V, Zubáčová Z, Treitli SC, Petrželková R, Eme L, Novák L, Žárský V, Barlow LD, Herman EK, Soukal P, Hroudová M, Doležal P, Stairs CW, Roger AJ, Eliáš M, Dacks JB, Vlček Č, Hampl V . A eukaryote without a mitochondrial organelle. Curr Biol, 2016,26(10):1274-1284.
doi: 10.1016/j.cub.2016.03.053 pmid: 27185558 |
| [89] |
Roger AJ, Muñoz-Gómez SA, Kamikawa R . The origin and diversification of mitochondria. Curr Biol, 2017,27(21):R1177-R1192.
doi: 10.1016/j.cub.2017.09.015 pmid: 29112874 |
| [90] |
López-García P, Eme L, Moreira D . Symbiosis in eukaryotic evolution. J Theor Biol, 2017,434:20-33.
doi: 10.1016/j.jtbi.2017.02.031 pmid: 28254477 |
| [91] |
Sagan L . On the origin of mitosing cells. J Theor Biol, 1967,14(3):255-274.
doi: 10.1016/0022-5193(67)90079-3 pmid: 11541392 |
| [92] |
Bonen L, Cunningham RS, Gray MW, Doolittle WF . Wheat embryo mitochondrial 18S ribosomal RNA: evidence for its prokaryotic nature. Nucleic Acids Res, 1977,4(3):663-671.
doi: 10.1093/nar/4.3.663 pmid: 866186 |
| [93] |
Schwartz RM, Dayhoff MO . Origins of prokaryotes, eukaryotes, mitochondria, and chloroplasts. Science, 1978,199(4327):395-403.
doi: 10.1126/science.202030 pmid: 202030 |
| [94] |
Yang D, Oyaizu Y, Oyaizu H, Olsen GJ, Woese CR . Mitochondrial origins. Proc Natl Acad Sci USA, 1985,82(13):4443-4447.
doi: 10.1073/pnas.82.13.4443 pmid: 3892535 |
| [95] |
Cavalier-Smith T . Eukaryotes with no mitochondria. Nature, 1987,326(6111):332-333.
doi: 10.1038/326332a0 pmid: 3561476 |
| [96] |
Cavalier-Smith T . Archaebacteria and archezoa. Nature, 1989,339(6220):100-101.
doi: 10.1038/339100a0 |
| [97] |
Cavalier-Smith T . The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int J Syst Evol Microbiol, 2002,52(2):297-354.
doi: 10.1099/00207713-52-2-297 |
| [98] |
Cavalier-Smith T . Origin of the cell nucleus, mitosis and sex: roles of intracellular coevolution. Biol Direct, 2010,5(1):7.
doi: 10.1186/1745-6150-5-7 |
| [99] |
Cavalier-Smith T . Only six kingdoms of life. Proc Biol Sci, 2004,271(1545):1251-1262.
doi: 10.1098/rspb.2004.2705 pmid: 15306349 |
| [100] |
Margulis L, Dolan MF, Whiteside JH . “Imperfections and oddities” in the origin of the nucleus. Paleobiology, 2005,31(2):175-191.
doi: 10.1666/0094-8373(2005)031[0175:IAOITO]2.0.CO;2 |
| [101] | Sapp J. Microbial Phylogeny and Evolution: Concepts and Controversies. Oxford: Oxford University Press on Demand, 2005, 184-206. |
| [102] |
Margulis L, Dolan MF, Guerrero R . The chimeric eukaryote: origin of the nucleus from the karyomastigont in amitochondriate protists. Proc Natl Acad Sci USA, 2000,97(13):6954-6959.
doi: 10.1073/pnas.97.13.6954 pmid: 10860956 |
| [103] |
Vellai T, Takács K, Vida G . A new aspect to the origin and evolution of eukaryotes. J Mol Evol, 1998,46(5):499-507.
doi: 10.1007/pl00006331 pmid: 9545461 |
| [104] | Searcy DG. Origins of mitochondria and chloroplasts from sulfur-based symbioses. In: Hartman H, Matsuno K eds. The Origin and Evolution of the Cell. Singapore: World Scientific, 1992, 47-78. |
| [105] | van der Giezen M, Tovar J, Clark CG . Mitochondrion- derived organelles in protists and fungi. Int Rev Cytol, 2005,244:175-225. |
| [106] |
Von Dohlen CD, Kohler S, Alsop ST, McManus WR. Mealybug β-proteobacterial endosymbionts contain γ-proteobacterial symbionts. Nature, 2001,412(6845):433-436.
doi: 10.1038/35086563 pmid: 11473316 |
| [107] |
Henze K, Martin W . How do mitochondrial genes get into the nucleus? Trends Genet, 2001,17(7):383-387.
doi: 10.1016/s0168-9525(01)02312-5 pmid: 11418217 |
| [108] |
Martin WF, Garg S, Zimorski V . Endosymbiotic theories for eukaryote origin. Philos Trans R Soc Lond B Biol Sci, 2015,370(1678):20140330.
pmid: 26323761 |
| [109] |
Lane N, Martin W . The energetics of genome complexity. Nature, 2010,467(7318):929-934.
doi: 10.1038/nature09486 pmid: 20962839 |
| [110] |
Garg SG, Martin WF . Mitochondria, the cell cycle, and the origin of sex via a syncytial eukaryote common ancestor. Genome Biol Evol, 2016,8(6):1950-1970.
doi: 10.1093/gbe/evw136 pmid: 27345956 |
| [111] |
Gould SB, Garg SG, Martin WF . Bacterial vesicle secretion and the evolutionary origin of the eukaryotic endomembrane system. Trends Microbiol, 2016,24(7):525-534.
doi: 10.1016/j.tim.2016.03.005 pmid: 27040918 |
| [112] |
McBride HM . Mitochondria and endomembrane origins. Curr Biol, 2018,28(8):R367-R372.
pmid: 29689215 |
| [113] |
Yutin N, Wolf MY, Wolf YI, Koonin EV . The origins of phagocytosis and eukaryogenesis. Biol Direct, 2009,4(1):9.
doi: 10.1186/1745-6150-4-9 |
| [114] |
Embley TM, Williams TA . Steps on the road to eukaryotes. Nature, 2015,521(7551):169-170.
doi: 10.1038/nature14522 pmid: 25945740 |
| [115] |
Burns JA, Pittis AA, Kim E . Gene-based predictive models of trophic modes suggest Asgard archaea are not phagocytotic. Nat Ecol Evol, 2018,2(4):697-704.
doi: 10.1038/s41559-018-0477-7 pmid: 29459706 |
| [116] |
Hampl V, Čepička I, Eliáš M . Was the mitochondrion necessary to start eukaryogenesis? Trends Microbiol, 2019,27(2):96-104.
doi: 10.1016/j.tim.2018.10.005 |
| [117] |
Pittis AA, Gabaldón T . Late acquisition of mitochondria by a host with chimaeric prokaryotic ancestry. Nature, 2016,531(7592):101-104.
doi: 10.1038/nature16941 pmid: 26840490 |
| [118] |
Ettema TJ . Evolution: Mitochondria in the second act. Nature, 2016,531(7592):39-40.
doi: 10.1038/nature16876 pmid: 26840482 |
| [119] |
Martin WF, Roettger M, Ku C, Garg SG, Nelson-Sathi S, Landan G . Late Mitochondrial origin is an artifact. Genome Biol Evol, 2017,9(2):373-379.
doi: 10.1093/gbe/evx027 pmid: 28199635 |
| [120] | Degli Esposti M. Phylogeny and Evolution of Bacteria and Mitochondria. CRC Press, 2018. |
| [121] |
Ku C, Nelson-Sathi S, Roettger M, Sousa FL, Lockhart PJ, Bryant D, Hazkani-Covo E, McInerney JO, Landan G, Martin WF. Endosymbiotic origin and differential loss of eukaryotic genes. Nature, 2015,524(7566):427-432.
doi: 10.1038/nature14963 pmid: 26287458 |
| [122] |
Andersson SG, Zomorodipour A, Andersson JO, Sicheritz-Pontén T, Alsmark UCM, Podowski RM, Näslund AK, Eriksson AS, Winkler HH, Kurland CG . The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature, 1998,396(6707):133-140.
doi: 10.1038/24094 pmid: 9823893 |
| [123] |
Fitzpatrick DA, Creevey CJ, McInerney JO. Genome phylogenies indicate a meaningful α-proteobacterial phylogeny and support a grouping of the mitochondria with the Rickettsiales. Mol Biol Evol, 2006,23(1):74-85.
doi: 10.1093/molbev/msj009 pmid: 16151187 |
| [124] |
Williams KP, Sobral BW, Dickerman AW . A robust species tree for the alphaproteobacteria. J Bacteriol, 2007,189(13):4578-4586.
doi: 10.1128/JB.00269-07 pmid: 17483224 |
| [125] |
Thrash JC, Boyd A, Huggett MJ, Grote J, Carini P, Yoder RJ, Robbertse B, Spatafora JW, Rappé MS, Giovannoni SJ . Phylogenomic evidence for a common ancestor of mitochondria and the SAR11 clade. Sci Rep, 2011,1:13.
pmid: 22355532 |
| [126] |
Brindefalk B, Ettema TJ, Viklund J, Thollesson M, Andersson SG . A phylometagenomic exploration of oceanic alphaproteobacteria reveals mitochondrial relatives unrelated to the SAR11 clade. PLoS One, 2011,6(9):e24457.
doi: 10.1371/journal.pone.0024457 pmid: 21935411 |
| [127] |
Rodríguez-Ezpeleta N, Embley TM . The SAR11 group of alpha-proteobacteria is not related to the origin of mitochondria. PLoS One, 2012,7(1):e30520.
doi: 10.1371/journal.pone.0030520 pmid: 22291975 |
| [128] |
Wang Z, Wu M . An integrated phylogenomic approach toward pinpointing the origin of mitochondria. Sci Rep, 2015,5:7949.
doi: 10.1038/srep07949 pmid: 25609566 |
| [129] |
Wang Z, Wu M . Phylogenomic reconstruction indicates mitochondrial ancestor was an energy parasite. PLoS One, 2014,9(10):e110685.
doi: 10.1371/journal.pone.0110685 pmid: 25333787 |
| [130] |
Martijn J, Vosseberg J, Guy L, Offre P, Ettema TJ . Deep mitochondrial origin outside the sampled alphaproteobacteria. Nature, 2018,557(7703):101-105.
pmid: 29695865 |
| [131] | Fan L, Wu DF, Goremykin V, Xiao J, Xu YB, Garg S, Zhang CL, Martin WF, Zhu RX . Mitochondria branch within Alphaproteobacteria. bioRxiv, 2019,715870. |
| [132] |
Le PT, Pontarotti P, Raoult D . Alphaproteobacteria species as a source and target of lateral sequence transfers. Trends Microbiol, 2014,22(3):147-156.
doi: 10.1016/j.tim.2013.12.006 |
| [133] |
Janouškovec J, Tikhonenkov DV, Burki F, Howe AT, Rohwer FL, Mylnikov AP, Keeling PJ. A new lineage of eukaryotes illuminates early mitochondrial genome reduction. Curr Biol, 2017, 27(23): 3717-3724.e5.
doi: 10.1016/j.cub.2017.10.051 pmid: 29174886 |
| [134] |
Muñoz-Gómez SA, Hess S, Burger G, Lang BF, Susko E, Slamovits CH, Roger AJ . An updated phylogeny of the Alphaproteobacteria reveals that the parasitic Rickettsiales and Holosporales have independent origins. eLife, 2019,8:e42535.
doi: 10.7554/eLife.42535 pmid: 30789345 |
| [135] |
Searcy DG . Metabolic integration during the evolutionary origin of mitochondria. Cell Res, 2003,13(4):229-238.
doi: 10.1038/sj.cr.7290168 pmid: 12974613 |
| [136] |
Lake JA, Rivera MC . Was the nucleus the first endosymbiont? Proc Natl Acad Sci USA, 1994,91(8):2880.
doi: 10.1073/pnas.91.8.2880 pmid: 8159671 |
| [137] |
Godde JS . Breaking through a phylogenetic impasse: a pair of associated archaea might have played host in the endosymbiotic origin of eukaryotes. Cell Biosci, 2012,2(1):29.
doi: 10.1186/2045-3701-2-29 pmid: 22913376 |
| [138] |
Forterre P, Gaïa M . Giant viruses and the origin of modern eukaryotes. Curr Opin Microbiol, 2016,31:44-49.
doi: 10.1016/j.mib.2016.02.001 pmid: 26894379 |
| [139] |
Bell PJL . The viral eukaryogenesis hypothesis: a key role for viruses in the emergence of eukaryotes from a prokaryotic world environment. Ann N Y Acad Sci, 2009,1178(1):91-105.
doi: 10.1111/j.1749-6632.2009.04994.x |
| [140] | Sousa FL, Neukirchen S, Allen JF, Lane N, Martin WF . Lokiarchaeon is hydrogen dependent. Nat Microbiol, 2016,1(5):1-3. |
| [141] |
Liu Y, Zhou ZC, Pan J, Baker BJ, Gu JD, Li M . Comparative genomic inference suggests mixotrophic lifestyle for Thorarchaeota. ISME J, 2018,12(4):1021-1031.
doi: 10.1038/s41396-018-0060-x pmid: 29445130 |
| [142] |
Manoharan L, Kozlowski JA, Murdoch RW, Löffler FE, Sousa FL, Schleper C . Metagenomes from coastal marine sediments give insights into the ecological role and cellular features of Loki-and Thorarchaeota. mBio, 2019,10(5):e02039-02019.
doi: 10.1128/mBio.02039-19 pmid: 31506313 |
| [143] |
Pushkarev A, Inoue K, Larom S, Flores-Uribe J, Singh M, Konno M, Tomida S, Ito S, Nakamura R, Tsunoda SP, Philosof A, Sharon I, Yutin N, Koonin EV, Kandori H, Béjà O . A distinct abundant group of microbial rhodopsins discovered using functional metagenomics. Nature, 2018,558(7711):595-599.
doi: 10.1038/s41586-018-0225-9 pmid: 29925949 |
| [144] |
Bulzu PA, Andrei AŞ, Salcher MM, Mehrshad M, Inoue K, Kandori H, Beja O, Ghai R, Banciu HL . Casting light on Asgardarchaeota metabolism in a sunlit microoxic niche. Nat Microbiol, 2019,4(7):1129-1137.
doi: 10.1038/s41564-019-0404-y pmid: 30936485 |
| [145] |
Spang A, Stairs CW, Dombrowski N, Eme L, Lombard J, Caceres EF, Greening C, Baker BJ, Ettema TJG . Proposal of the reverse flow model for the origin of the eukaryotic cell based on comparative analyses of Asgard archaeal metabolism. Nat Microbiol, 2019,4(7):1138-1148.
doi: 10.1038/s41564-019-0406-9 |
| [146] |
Saghaï A, Gutiérrez-Preciado A, Deschamps P, Moreira D, Bertolino P, Ragon M, López-García P . Unveiling microbial interactions in stratified mat communities from a warm saline shallow pond. Environ Microbiol, 2017,19(6):2405-2421.
doi: 10.1111/1462-2920.13754 pmid: 28489281 |
| [147] |
Akıl C, Robinson RC . Genomes of Asgard archaea encode profilins that regulate actin. Nature, 2018,562(7727):439-443.
doi: 10.1038/s41586-018-0548-6 pmid: 30283132 |
| [148] |
Imachi H, Aoi K, Tasumi E, Saito Y, Yamanaka Y, Saito Y, Yamaguchi T, Tomaru H, Takeuchi R, Morono Y, Inagaki F, Takai K . Cultivation of methanogenic community from subseafloor sediments using a continuous- flow bioreactor. ISME J, 2011,5(12):1913-1925.
doi: 10.1038/ismej.2011.64 |
| [149] |
Szöllősi GJ, Tannier E, Daubin V, Boussau B . The inference of gene trees with species trees. Syst Biol, 2015,64(1):e42-e62.
doi: 10.1093/sysbio/syu048 pmid: 25070970 |
| [150] |
Hu GB . Whole cell cryo-electron tomography suggests mitochondria divide by budding. Microsc Microanal, 2014,20(4):1180-1187.
doi: 10.1017/S1431927614001317 |
| [151] |
Mehta AP, Supekova L, Chen JH, Pestonjamasp K, Webster P, Ko Y, Henderson SC, McDermott G, Supek F, Schultz PG. Engineering yeast endosymbionts as a step toward the evolution of mitochondria. Proc Natl Acad Sci USA, 2018,115(46):11796-11801.
doi: 10.1073/pnas.1813143115 pmid: 30373839 |
| [1] | 宋睿嘉, 韩露, 孙海峰, 沈彬. 线粒体DNA碱基编辑技术研究进展[J]. 遗传, 2023, 45(8): 632-642. |
| [2] | 张茜, 王子豪, 田烨. 跨组织线粒体应激信号交流调控机体衰老研究进展[J]. 遗传, 2023, 45(3): 187-197. |
| [3] | 张爽, 郭珊珊, 王汝雯, 马仁燕, 吴显敏, 陈佩杰, 王茹. PARK基因家族在骨骼肌肌病中的研究进展[J]. 遗传, 2022, 44(7): 545-555. |
| [4] | 袁洁, 蔡时青. 衰老过程中行为和认知功能退化的调控机制研究[J]. 遗传, 2021, 43(6): 545-570. |
| [5] | 吴安平, 庆宏, 全贞贞. Rab蛋白家族在神经类疾病中的作用[J]. 遗传, 2021, 43(1): 16-29. |
| [6] | 谌阳, 王文君, 付明, 徐国强, 周翔, 刘榜. 基于核质遗传原理建立多重PCR检测方法鉴定阿胶中马、驴源性成分及皮张种源[J]. 遗传, 2020, 42(10): 1028-1035. |
| [7] | 刘传明,丁利军,李佳音,戴建武,孙海翔. 衰老导致卵巢功能低下研究进展[J]. 遗传, 2019, 41(9): 816-826. |
| [8] | 匡卫民, 于黎. 基因组时代线粒体基因组拼装策略及软件应用现状[J]. 遗传, 2019, 41(11): 979-993. |
| [9] | 潘云枫, 王演怡, 陈静雯, 范怡梅. 线粒体代谢介导的表观遗传改变与衰老研究[J]. 遗传, 2019, 41(10): 893-904. |
| [10] | 孙吉吉, 赵晓旭, 乔丽华, 梅霜, 聂志鹏, 张青海, 冀延春, 蒋萍萍, 管敏鑫. 线粒体遗传疾病细胞模型的构建:永生淋巴细胞系和转线粒体细胞系[J]. 遗传, 2016, 38(7): 666-673. |
| [11] | 贾振伟. 线粒体与多潜能干细胞功能[J]. 遗传, 2016, 38(7): 603-611. |
| [12] | 李雪娟, 黄原, 雷富民. 山鹧鸪属鸟类线粒体基因组的比较及系统发育研究[J]. 遗传, 2014, 36(9): 912-920. |
| [13] | 孟祥娟, 朱金萍, 高敏, 张赛, 赵福新, 张娟娟, 刘晓玲, 韦企平, 童绎, 张铭连, 瞿佳, 管敏鑫. 中国人群携带m.14484T>C突变的Leber’s遗传性视神经病变线粒体单体型及多态位点分析[J]. 遗传, 2014, 36(4): 336-345. |
| [14] | 许美芬, 何轶群, 耿军伟, 孟燕子, 于涵, 林枝, 施苏雪, 薛凌, 卢中秋, 管敏鑫. 两个携带线粒体tRNAMet/tRNAGlnA4401G和tRNACysG5821A突变的中国汉族原发性高血压家系的临床及分子遗传学特征[J]. 遗传, 2014, 36(2): 127-134. |
| [15] | 刘莉, 邵宇权, 张宝荣, 蒋萍萍, 都爱莲, 管敏鑫. 6个线粒体脑肌病伴高乳酸血症和卒中样发作综合征(MELAS)家系先证者线粒体基因全序列比较分析[J]. 遗传, 2014, 36(11): 1159-1167. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||

www.chinagene.cn
备案号:京ICP备09063187号-4
总访问:,今日访问:,当前在线:

