遗传 ›› 2021, Vol. 43 ›› Issue (9): 858-879.doi: 10.16288/j.yczz.21-170
王雅楠1( ), 徐涛2(
), 徐涛2( ), 王万鹏1, 张庆祝1,3(
), 王万鹏1, 张庆祝1,3( ), 解莉楠1,4(
), 解莉楠1,4( )
)
收稿日期:2021-05-11
修回日期:2021-07-14
出版日期:2021-09-20
发布日期:2021-09-03
通讯作者:
张庆祝,解莉楠
E-mail:yn-wang@foxmail.com;tao.xv@nefu.edu.cn;qingzhu.zhang@nefu.edu.cn;linanxie@nefu.edu.cn
作者简介:王雅楠,本科,专业方向:生物科学。E-mail: 基金资助:
Wang Ya'nan1( ), Tao Xu2(
), Tao Xu2( ), Wanpeng Wang1, Qingzhu Zhang1,3(
), Wanpeng Wang1, Qingzhu Zhang1,3( ), Xie Li'nan1,4(
), Xie Li'nan1,4( )
)
Received:2021-05-11
Revised:2021-07-14
Online:2021-09-20
Published:2021-09-03
Contact:
Zhang Qingzhu,Xie Li'nan
E-mail:yn-wang@foxmail.com;tao.xv@nefu.edu.cn;qingzhu.zhang@nefu.edu.cn;linanxie@nefu.edu.cn
Supported by:摘要:
表观遗传修饰是指染色体DNA和组蛋白上的化学修饰,主要包括DNA甲基化、组蛋白修饰和非编码RNA。在不改变DNA序列的情况下,这些修饰可以通过改变染色质状态来影响遗传信息的表达,并具有可遗传性,对植物的生长发育具有重要的调控作用。当特定的表观遗传修饰发生改变时,农作物可以获得优异的表型、更强的环境适应性,因此人为改变表观遗传修饰有望获得更适于农业生产的优质种质资源。本文综述了植物表观遗传修饰的主要类型及其在作物重要性状的形成和环境胁迫响应中的相关研究进展,总结了表观遗传学应用于作物改良必须解决的主要问题,以期在表观遗传修饰层面为作物的育种改良提供新思路。
王雅楠, 徐涛, 王万鹏, 张庆祝, 解莉楠. 表观遗传修饰在作物重要性状形成中的作用[J]. 遗传, 2021, 43(9): 858-879.
Wang Ya'nan, Tao Xu, Wanpeng Wang, Qingzhu Zhang, Xie Li'nan. Role of epigenetic modifications in the development of crops essential traits[J]. Hereditas(Beijing), 2021, 43(9): 858-879.
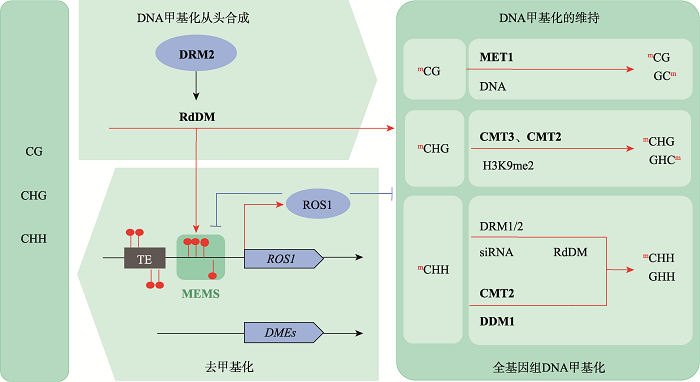
图1
拟南芥DNA甲基化的从头合成、维持和去甲基化 图根据参考文献[4]修改绘制。DRM2:DOMAINS REARRANGED METHYLTRANSFERASE 2,甲基化转移酶2;RdDM:RNA directed DNA methylation,RNA介导的DNA甲基化;MEMS:methylation monitoring sequence,甲基化监测序列;MET1: METHYLTRANSFERASE 1,甲基转移酶1;CMT3:CHROMOMETHYLASE 3,甲基转移酶3;CMT2:CHROMOMETHYLASE 2,甲基转移酶2;DDM1:DECREASED DNA METHYLATION 1,染色质重塑因子。基因上的红色圆圈标记代表DNA甲基化修饰;DNA主动去甲基化过程主要涉及2类主要的DNA去甲基化酶—ROS1和DMEs。"
| [1] | Jiang J, Qian Q, Ma BJ, Gao ZY. Epigenetic variation and its application in crop improvement. Hereditas (Beijing), 2014, 36(5):469-475. |
| 江静, 钱前, 马伯军, 高振宇. 表观遗传变异及其在作物改良中的应用. 遗传, 2014, 36(5):469-475. | |
| [2] |
Zhang PY, Wang JG, Geng YP, Dai JR, Zhong Y, Chen ZZ, Zhu K, Wang XZ, Chen SY. MSAP-based analysis of DNA methylation diversity in tobacco exposed to different environments and at different development phases. Biochem Syst Ecol, 2015, 62:249-260.
doi: 10.1016/j.bse.2015.09.009 |
| [3] |
Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet, 2010, 11(3):204-220.
doi: 10.1038/nrg2719 |
| [4] |
Zhang HM, Lang ZB, Zhu JK. Dynamics and function of DNA methylation in plants. Nat Rev Mol Cell Biol, 2018, 19(8):489-506.
doi: 10.1038/s41580-018-0016-z |
| [5] |
Tirnaz S, Batley J. DNA methylation: toward crop disease resistance improvement. Trends Plant Sci, 2019, 24(12):1137-1150.
doi: 10.1016/j.tplants.2019.08.007 |
| [6] |
Lei MG, Zhang HM, Julian R, Tang K, Xie SJ, Zhu JK. Regulatory link between DNA methylation and active demethylation in Arabidopsis. Proc Natl Acad Sci USA, 2015, 112(11):3553-3557.
doi: 10.1073/pnas.1502279112 |
| [7] |
Aquino EM, Benton MC, Haupt LM, Sutherland HG, Griffiths LR, Lea RA. Current understanding of DNA methylation and age-related disease. OBM Genet, 2018, 2(2): doi: 10.21926/obm.genet.1802016.
doi: 10.21926/obm.genet.1802016 |
| [8] |
Soppe WJ, Jacobsen SE, Alonso-Blanco C, Jackson JP, Kakutani T, Koornneef M, Peeters AJ. The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol Cell, 2000, 6(4):791-802.
pmid: 11090618 |
| [9] |
Jiao YL, Deng XW. A genome-wide transcriptional activity survey of rice transposable element-related genes. Genome Biol, 2007, 8(2):R28.
doi: 10.1186/gb-2007-8-2-r28 |
| [10] |
Schnable PS, Ware D, Fulton RS, Stein JC, Wei FS, Pasternak S, Liang CZ, Zhang JW, Fulton L, Graves TA, Minx P, Reily AD, Courtney L, Kruchowski SS, Tomlinson C, Strong C, Delehaunty K, Fronick C, Courtney B, Rock SM, Belter E, Du FY, Kim K, Abbott RM, Cotton M, Levy A, Marchetto P, Ochoa K, Jackson SM, Gillam B, Chen WZ, Yan L, Higginbotham J, Cardenas M, Waligorski J, Applebaum E, Phelps L, Falcone J, Kanchi K, Thane T, Scimone A, Thane N, Henke J, Wang T, Ruppert J, Shah N, Rotter K, Hodges J, Ingenthron E, Cordes M, Kohlberg S, Sgro J, Delgado B, Mead K, Chinwalla A, Leonard S, Crouse K, Collura K, Kudrna D, Currie J, He RF, Angelova A, Rajasekar S, Mueller T, Lomeli R, Scara G, Ko A, Delaney K, Wissotski M, Lopez G, Campos D, Braidotti M, Ashley E, Golser W, Kim H, Lee S, Lin JK, Dujmic Z, Kim W, Talag J, Zuccolo A, Fan CZ, Sebastian A, Kramer M, Spiegel L, Nascimento L, Zutavern T, Miller B, Ambroise C, Muller S, Spooner W, Narechania A, Ren LY, Wei S, Kumari S, Faga B, Levy MJ, McMahan L, Van Buren P, Vaughn MW, Ying K, Yeh CT, Emrich SJ, Jia Y, Kalyanaraman A, Hsia AP, Barbazuk WB, Baucom RS, Brutnell TP, Carpita NC, Chaparro C, Chia JM, Deragon JM, Estill JC, Fu Y, Jeddeloh JA, Han YJ, Lee H, Li PH, Lisch DR, Liu SZ, Liu ZJ, Nagel DH, McCann MC, SanMiguel P, Myers AM, Nettleton D, Nguyen J, Penning BW, Ponnala L, Schneider KL, Schwartz DC, Sharma A, Soderlund C, Springer NM, Sun Q, Wang H, Waterman M, Westerman R, Wolfgruber TK, Yang LX, Yu Y, Zhang LF, Zhou SG, Zhu QH, Bennetzen JL, Dawe RK, Jiang JM, Jiang N, Presting GG, Wessler SR, Aluru S, Martienssen RA, Clifton SW, McCombie WR, Wing RA, Wilson RK. The B73 maize genome: complexity, diversity, and dynamics. Science, 2009, 326(5956):1112-1115.
doi: 10.1126/science.1178534 pmid: 19965430 |
| [11] |
Zemach A, Kim MY, Silva P, Rodrigues JA, Dotson B, Brooks MD, Zilberman D. Local DNA hypomethylation activates genes in rice endosperm. Proc Natl Acad Sci USA, 2010, 107(43):18729-18734.
doi: 10.1073/pnas.1009695107 |
| [12] |
Zemach A, Mcdaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science, 2010, 328(5980):916-919.
doi: 10.1126/science.1186366 pmid: 20395474 |
| [13] |
Cho J. Transposon-derived non-coding RNAs and their function in plants. Front Plant Sci, 2018, 9:600.
doi: 10.3389/fpls.2018.00600 |
| [14] |
Perrone A, Martinelli F. Plant stress biology in epigenomic era. Plant Sci, 2020, 294:110376.
doi: S0168-9452(19)31549-3 pmid: 32234231 |
| [15] |
Matsunaga S. Junk DNA promotes sex chromosome evolution. Heredity (Edinb), 2009, 102(6):525-526.
doi: 10.1038/hdy.2009.36 pmid: 19337304 |
| [16] |
Ito H, Yoshida T, Tsukahara S, Kawabe A. Evolution of theONSEN retrotransposon family activated upon heat stress in Brassicaceae. Gene, 2013, 518(2):256-261.
doi: 10.1016/j.gene.2013.01.034 |
| [17] |
Ito H, Kim JM, Matsunaga W, Saze H, Matsui A, Endo TA, Harukawa Y, Takagi H, Yaegashi H, Masuta Y, Masuda S, Ishida J, Tanaka M, Takahashi S, Morosawa T, Toyoda T, Kakutani T, Kato A, Seki M. A stress- activated transposon in Arabidopsis induces transgenerational abscisic acid insensitivity. Sci Rep, 2016, 6:23181.
doi: 10.1038/srep23181 |
| [18] |
Schmitz RJ, Schultz MD, Urich MA, Nery JR, Pelizzola M, Libiger O, Alix A, Mccosh RB, Chen HM, Schork NJ, Ecker JR. Patterns of population epigenomic diversity. Nature, 2013, 495(7440):193-198.
doi: 10.1038/nature11968 |
| [19] |
Wang XT, Zhang ZB, Fu TS, Hu LJ, Xu CM, Gong L, Wendel JF, Liu B. Gene-body CG methylation and divergent expression of duplicate genes in rice. Sci Rep, 2017, 7(1):2675.
doi: 10.1038/s41598-017-02860-4 |
| [20] |
Do Kim K, El Baidouri M, Abernathy B, Iwata-Otsubo A, Chavarro C, Gonzales M, Libault M, Grimwood J, Jackson SA. A comparative epigenomic analysis of polyploidy-derived genes in soybean and common bean. Plant Physiol, 2015, 168(4):1433-1447.
doi: 10.1104/pp.15.00408 |
| [21] |
González RM, Ricardi MM, Iusem ND. Atypical epigenetic mark in an atypical location: cytosine methylation at asymmetric (CNN) sites within the body of a non-repetitive tomato gene. BMC Plant Biol, 2011, 11(1):94.
doi: 10.1186/1471-2229-11-94 pmid: 21599976 |
| [22] |
Richard MMS, Gratias A, Thareau V, Do Kim K, Balzergue S, Joets J, Jackson SA, Geffroy V. Genomic and epigenomic immunity in common bean: the unusual features of NB-LRR gene family. DNA Res, 2018, 25(2):161-172.
doi: 10.1093/dnares/dsx046 pmid: 29149287 |
| [23] |
Peterson CL, Laniel MA. Histones and histone modifications. Curr Biol, 2004, 14(14):R546-R551.
doi: 10.1016/j.cub.2004.07.007 |
| [24] |
Jambhekar A, Dhall A, Shi Y. Roles and regulation of histone methylation in animal development. Nat Rev Mol Cell Biol, 2019, 20(10):625-641.
doi: 10.1038/s41580-019-0151-1 |
| [25] | Jiang XX, Li XY, Ma Y, Müller-Xing R, Xing Q. Research progress of epigenetic regulatory gene CLF in morphogenesis of plant organs. Soyb Bull, 2020, 3:(3): 33-39, 45. |
| 姜晓旭, 李晓屿, 马瑶, Müller-Xing R, 邢倩. 表观遗传调控基因CLF在植物器官形态建成的研究进展. 大豆科技, 2020, (3):33-39, 45. | |
| [26] |
Lopez-Vernaza M, Yang SX, Muller R, Thorpe F, de Leau E, Goodrich J. Antagonistic roles ofSEPALLATA3, FT and FLC genes as targets of the polycomb group gene CURLY LEAF. PLoS One, 2012, 7(2):e30715.
doi: 10.1371/journal.pone.0030715 |
| [27] |
Bian SM, Li J, Tian G, Cui YH, Hou YM, Qiu WD. Combinatorial regulation of CLF and SDG8 during Arabidopsis shoot branching. Acta Physiol Plant, 2016, 38(7):1-11.
doi: 10.1007/s11738-015-2023-4 |
| [28] |
Mandel T, Moreau F, Kutsher Y, Fletcher JC, Carles CC, Eshed Williams L. The ERECTA receptor kinase regulates Arabidopsis shoot apical meristem size, phyllotaxy and floral meristem identity. Development, 2014, 141(4):830-841.
doi: 10.1242/dev.104687 pmid: 24496620 |
| [29] |
Park EY, Tsuyuki KM, Hu FL, Lee J, Jeong J. PRC2- mediated H3K27me3 contributes to transcriptional regulation of FIT-dependent iron deficiency response. Front Plant Sci, 2019, 10:627.
doi: 10.3389/fpls.2019.00627 |
| [30] |
Zhang XY, Bernatavichute YV, Cokus S, Pellegrini M, Jacobsen SE. Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol, 2009, 10(6):R62.
doi: 10.1186/gb-2009-10-6-r62 |
| [31] |
Minow MAA, Colasanti J. Does variable epigenetic inheritance fuel plant evolution? Genome, 2020, 63(5):253-262.
doi: 10.1139/gen-2019-0190 pmid: 32053387 |
| [32] |
Vignali M, Hassan AH, Neely KE, Workman JL. ATP-dependent chromatin-remodeling complexes. Mol Cell Biol, 2000, 20(6):1899-1910.
doi: 10.1128/MCB.20.6.1899-1910.2000 pmid: 10688638 |
| [33] |
Hota SK, Bruneau BG. ATP-dependent chromatin remodeling during mammalian development. Development, 2016, 143(16):2882-2897.
doi: 10.1242/dev.128892 |
| [34] |
Mlynárová L, Nap JP, Bisseling T. The SWI/SNF chromatin-remodeling geneAtCHR12 mediates temporary growth arrest in Arabidopsis thaliana upon perceiving environmental stress. Plant J, 2007, 51(5):874-885.
pmid: 17605754 |
| [35] | Wang RX, Xu JH. Genomic DNA methylation and histone methylation. Hereditas (Beijing), 2014, 36(3):191-199. |
| 王瑞娴, 徐建红. 基因组DNA甲基化及组蛋白甲基化. 遗传, 2014, 36(3):191-199. | |
| [36] | Zhang SF, Li XR, Sun CB, He YK. Epigenetics of plant vernalization regulated by non-coding RNAs. Hereditas (Beijing), 2012, 34(7):829-834. |
| 张绍峰, 李晓荣, 孙传宝, 何玉科. 植物非编码RNA调控春化作用的表观遗传. 遗传, 2012, 34(7):829-834. | |
| [37] |
Pajoro A, Severing E, Angenent GC, Immink RGH. Histone H3 lysine 36 methylation affects temperature- induced alternative splicing and flowering in plants. Genome Biol, 2017, 18(1):102.
doi: 10.1186/s13059-017-1235-x pmid: 28566089 |
| [38] |
Kim SY, Park BS, Kwon SJ, Kim J, Lim MH, Park YD, Kim DY, Suh SC, Jin YM, Ahn JH, Lee YH. Delayed flowering time in Arabidopsis and Brassica rapa by the overexpression of FLOWERING LOCUS C (FLC) homologs isolated from Chinese cabbage(Brassica rapa L.: ssp. pekinensis). Plant Cell Rep, 2007, 26(3):327-336.
doi: 10.1007/s00299-006-0243-1 |
| [39] |
Lin SI, Wang JG, Poon SY, Su CL, Wang SS, Chiou TJ. Differential regulation of FLOWERING LOCUS C expression by vernalization in cabbage andArabidopsis. Plant Physiol, 2005, 137(3):1037-1048.
doi: 10.1104/pp.104.058974 |
| [40] | Shen X, Chen XH, Xu XP, Huo W, Li XF, Jiang MQ, Zhang J, Lin YL, Lai ZX. Genome-wide identification and expression analysis of SDG gene family during early somatic embryogenesis inDimocarpus longan Lour. Chin J of Trop Crop, 2019, 40(10):1889-1901. |
| 申序, 陈晓慧, 徐小萍, 霍雯, 李晓斐, 蒋梦琦, 张婧, 林玉玲, 赖钟雄. 龙眼体细胞胚胎发生早期SDG基因家族的全基因组鉴定与表达分析. 热带作物学报, 2019, 40(10):1889-1901. | |
| [41] |
Shafiq S, Berr A, Shen WH. Combinatorial functions of diverse histone methylations inArabidopsis thaliana flowering time regulation. New Phytol, 2014, 201(1):312-322.
doi: 10.1111/nph.2013.201.issue-1 |
| [42] |
Liu KP, Yu Y, Dong AW, Shen WH. SET DOMAIN GROUP701 encodes a H3K4-methytransferase and regulates multiple key processes of rice plant development. New Phytol, 2017, 215(2):609-623.
doi: 10.1111/nph.2017.215.issue-2 |
| [43] |
Springer NM. Epigenetics and crop improvement. Trends Genet, 2013, 29(4):241-247.
doi: 10.1016/j.tig.2012.10.009 pmid: 23128009 |
| [44] |
Liu B, Wei G, Shi JL, Jin J, Shen T, Ni T, Shen WH, Yu Y, Dong AW. SET DOMAIN GROUP 708, a histone H3 lysine 36-specific methyltransferase, controls flowering time in rice (Oryza sativa). New Phytol, 2016, 210(2):577-588.
doi: 10.1111/nph.2016.210.issue-2 |
| [45] |
Sang Q, Pajoro A, Sun HQ, Song BX, Yang X, Stolze SC, Andrés F, Schneeberger K, Nakagami H, Coupland G. Mutagenesis of a quintuple mutant impaired in environmental responses reveals roles forCHROMATIN REMODELING4 in the Arabidopsis floral transition. Plant Cell, 2020, 32(5):1479-1500.
doi: 10.1105/tpc.19.00992 |
| [46] |
Teyssier E, Bernacchia G, Maury S, How Kit A, Stammitti-Bert L, Rolin D, Gallusci P. Tissue dependent variations of DNA methylation and endoreduplication levels during tomato fruit development and ripening. Planta, 2008, 228(3):391-399.
doi: 10.1007/s00425-008-0743-z pmid: 18488247 |
| [47] | Shen Y, Conde E Silva N, Audonnet L, Servet C, Wei W, Zhou DX. Over-expression of histone H3K4 demethylase gene JMJ15 enhances salt tolerance in Arabidopsis. Front Plant Sci, 2014, 5:290. |
| [48] |
Yao MQ, Chen WW, Kong JH, Zhang XL, Shi NN, Zhong SL, Ma P, Gallusci P, Jackson S, Liu YL, Hong YG. METHYLTRANSFERASE1 and ripening modulate vivipary during tomato fruit development. Plant Physiol, 2020, 183(4):1883-1897.
doi: 10.1104/pp.20.00499 |
| [49] |
Liu RE, How-Kit A, Stammitti L, Teyssier E, Rolin D, Mortain-Bertrand A, Halle S, Liu MC, Kong JH, Wu CQ, Degraeve-Guibault C, Chapman NH, Maucourt M, Hodgman TC, Tost J, Bouzayen M, Hong YG, Seymour GB, Giovannoni JJ, Gallusci P. A DEMETER-like DNA demethylase governs tomato fruit ripening. Proc Natl Acad Sci USA, 2015, 112(34):10804-10809.
doi: 10.1073/pnas.1503362112 |
| [50] |
Lang ZB, Wang YH, Tang K, Tang DG, Datsenka T, Cheng JF, Zhang YJ, Handa AK, Zhu JK. Critical roles of DNA demethylation in the activation of ripening- induced genes and inhibition of ripening-repressed genes in tomato fruit. Proc Natl Acad Sci USA, 2017, 114(22):E4511-E4519.
doi: 10.1073/pnas.1705233114 |
| [51] |
Huang H, Liu RE, Niu QF, Tang K, Zhang B, Zhang H, Chen KS, Zhu JK, Lang ZB. Global increase in DNA methylation during orange fruit development and ripening. Proc Natl Acad Sci USA, 2019, 116(4):1430-1436.
doi: 10.1073/pnas.1815441116 |
| [52] |
Cheng JF, Niu QF, Zhang B, Chen KS, Yang RH, Zhu JK, Zhang YJ, Lang ZB. Downregulation of RdDM during strawberry fruit ripening. Genome Biol, 2018, 19(1):212.
doi: 10.1186/s13059-018-1587-x |
| [53] |
Quadrana L, Almeida J, Asís R, Duffy T, Dominguez PG, Bermúdez L, Conti G, Corrêa da Silva JV, Peralta IE, Colot V, Asurmendi S, Fernie AR, Rossi M, Carrari F. Natural occurring epialleles determine vitamin E accumulation in tomato fruits. Nat Commun, 2014, 5:3027.
doi: 10.1038/ncomms5027 pmid: 24967512 |
| [54] |
Guo JE, Hu ZL, Zhu MK, Li FF, Zhu ZG, Lu Y, Chen GP. The tomato histone deacetylase SlHDA1 contributes to the repression of fruit ripening and carotenoid accumulation. Sci Rep, 2017, 7(1):7930.
doi: 10.1038/s41598-017-08512-x |
| [55] | Han YC, Kuang JF, Chen JY, Liu XC, Xiao YY, Fu CC, Wang JN, Wu KQ, Lu WJ. Banana transcription factor MaERF11 recruits histone deacetylase MaHDA1 and represses the expression of MaACO1 and expansins during fruit ripening. Plant Physiol, 2016, 171(2):1070-1084. |
| [56] |
Li ZW, Jiang GX, Liu XC, Ding XC, Zhang DD, Wang XW, Zhou YJ, Yan HL, Li TT, Wu KQ, Jiang YM, Duan XW. Histone demethylase SlJMJ6 promotes fruit ripening by removing H3K27 methylation of ripening- related genes in tomato. New Phytol, 2020, 227(4):1138-1156.
doi: 10.1111/nph.v227.4 |
| [57] |
Nie WF, Lei MG, Zhang MX, Tang K, Huang H, Zhang CJ, Miki D, Liu P, Yang Y, Wang XG, Zhang H, Lang ZB, Liu N, Xu XC, Yelagandula R, Zhang HM, Wang ZD, Chai XQ, Andreucci A, Yu JQ, Berger F, Lozano- Duran R, Zhu JK. Histone acetylation recruits the SWR1 complex to regulate active DNA demethylation inArabidopsis. Proc Natl Acad Sci USA, 2019, 116(33):16641-16650.
doi: 10.1073/pnas.1906023116 |
| [58] |
Yelagandula R, Stroud H, Holec S, Zhou KD, Feng SH, Zhong XH, Muthurajan UM, Nie X, Kawashima T, Groth M, Luger K, Jacobsen SE, Berger F. The histone variant H2A.W defines heterochromatin and promotes chromatin condensation inArabidopsis. Cell, 2014, 158(1):98-109.
doi: 10.1016/j.cell.2014.06.006 pmid: 24995981 |
| [59] |
Yang XD, Zhang XL, Yang YX, Zhang H, Zhu WM, Nie WF. The histone variant Sl_H2A.Z regulates carotenoid biosynthesis and gene expression during tomato fruit ripening. Hortic Res, 2021, 8(1):85.
doi: 10.1038/s41438-021-00520-3 |
| [60] | Gallusci P, Hodgman C, Teyssier E, Seymour GB. DNA methylation and chromatin regulation during fleshy fruit development and ripening. Front Plant Sci, 2016, 7:807. |
| [61] |
Zhang XQ, Sun J, Cao XF, Song XW. Epigenetic mutation of RAV6 affects leaf angle and seed size in rice. Plant Physiol, 2015, 169(3):2118-2128.
doi: 10.1104/pp.15.00836 |
| [62] |
Xu L, Yuan K, Yuan M, Meng XB, Chen M, Wu JG, Li JY, Qi YJ. Regulation of rice tillering by RNA-directed DNA methylation at miniature inverted-repeat transposable elements. Mol Plant, 2020, 13(6):851-863.
doi: 10.1016/j.molp.2020.02.009 |
| [63] |
Chen M, Lin JY, Hur J, Pelletier JM, Baden R, Pellegrini M, Harada JJ, Goldberg RB. Seed genome hypomethylated regions are enriched in transcription factor genes. Proc Natl Acad Sci USA, 2018, 115(35):E8315-E8322.
doi: 10.1073/pnas.1811017115 |
| [64] |
Lin JY, Le BH, Chen M, Henry KF, Hur J, Hsieh TF, Chen PY, Pelletier JM, Pellegrini M, Fischer RL, Harada JJ, Goldberg RB. Similarity between soybean and Arabidopsis seed methylomes and loss of non-CG methylation does not affect seed development. Proc Natl Acad Sci USA, 2017, 114(45):E9730-E9739.
doi: 10.1073/pnas.1716758114 |
| [65] |
Liu JX, Wu XB, Yao XF, Yu R, Larkin PJ, Liu CM. Mutations in the DNA demethylase OsROS1 result in a thickened aleurone and improved nutritional value in rice grains. Proc Natl Acad Sci USA, 2018, 115(44):11327-11332.
doi: 10.1073/pnas.1806304115 |
| [66] |
Rajkumar MS, Gupta K, Khemka NK, Garg R, Jain M. DNA methylation reprogramming during seed development and its functional relevance in seed size/weight determination in chickpea. Commun Biol, 2020, 3(1):340.
doi: 10.1038/s42003-020-1059-1 pmid: 32620865 |
| [67] |
Telias A, Lin-Wang K, Stevenson DE, Cooney JM, Hellens RP, Allan AC, Hoover EE, Bradeen JM. Apple skin patterning is associated with differential expression ofMYB10. BMC Plant Biol, 2011, 11:93.
doi: 10.1186/1471-2229-11-93 |
| [68] |
Qian MJ, Sun YW, Allan AC, Teng YW, Zhang D. The red sport of ‘Zaosu’ pear and its red-striped pigmentation pattern are associated with demethylation of thePyMYB10 promoter. Phytochemistry, 2014, 107:16-23.
doi: 10.1016/j.phytochem.2014.08.001 |
| [69] |
Ong-Abdullah M, Ordway JM, Jiang N, Ooi SE, Kok SY, Sarpan N, Azimi N, Hashim AT, Ishak Z, Rosli SK, Malike FA, Bakar NAA, Marjuni M, Abdullah N, Yaakub Z, Amiruddin MD, Nookiah R, Singh R, Low ETL, Chan KL, Azizi N, Smith SW, Bacher B, Budiman MA, Van Brunt A, Wischmeyer C, Beil M, Hogan M, Lakey N, Lim CC, Arulandoo X, Wong CK, Choo CN, Wong WC, Kwan YY, Alwee SSRS, Sambanthamurthi R, Martienssen RA. Loss of Karma transposon methylation underlies the mantled somaclonal variant of oil palm. Nature, 2015, 525(7570):533-537.
doi: 10.1038/nature15365 |
| [70] |
Raju SKK, Shao MR, Sanchez R, Xu YZ, Sandhu A, Graef G, Mackenzie S. An epigenetic breeding system in soybean for increased yield and stability. Plant Biotechnol J, 2018, 16(11):1836-1847.
doi: 10.1111/pbi.2018.16.issue-11 |
| [71] | Liu J, Deng SL, Wang H, Ye J, Wu HW, Sun HX, Chua NH. CURLY LEAF regulates gene sets coordinating seed size and lipid biosynthesis in Arabidopsis. Plant Physiol, 2016, 171(1):424-436. |
| [72] |
Tricker PJ. Transgenerational inheritance or resetting of stress-induced epigenetic modifications: two sides of the same coin. Front Plant Sci, 2015, 6:699.
doi: 10.3389/fpls.2015.00699 pmid: 26442015 |
| [73] |
Liu X, Luo M, Zhang W, Zhao JH, Zhang JX, Wu KQ, Tian LN, Duan J. Histone acetyltransferases in rice (Oryza sativa L.): phylogenetic analysis, subcellular localization and expression. BMC Plant Biol, 2012, 12:145.
doi: 10.1186/1471-2229-12-145 |
| [74] |
Akimoto K, Katakami H, Kim HJ, Ogawa E, Sano CM, Wada Y, Sano H. Epigenetic inheritance in rice plants. Ann Bot, 2007, 100(2):205-217.
doi: 10.1093/aob/mcm110 |
| [75] |
Ding B, Bellizzi MDR, Ning Y, Meyers BC, Wang GL. HDT701, a histone H4 deacetylase, negatively regulates plant innate immunity by modulating histone H4 acetylation of defense-related genes in rice. Plant Cell, 2012, 24(9):3783-3794.
doi: 10.1105/tpc.112.101972 |
| [76] |
Hou YX, Wang LY, Wang L, Liu LM, Li L, Sun L, Rao Q, Zhang J, Huang SW. JMJ704 positively regulates rice defense response againstXanthomonas oryzae pv. oryzae infection via reducing H3K4me2/3 associated with negative disease resistance regulators. BMC Plant Biol, 2015, 15:286.
doi: 10.1186/s12870-015-0674-3 |
| [77] |
Luo X, He YH. Experiencing winter for spring flowering: A molecular epigenetic perspective on vernalization. J Integr Plant Biol, 2020, 62(1):104-117.
doi: 10.1111/jipb.v62.1 |
| [78] |
Zuther E, Schaarschmidt S, Fischer A, Erban A, Pagter M, Mubeen U, Giavalisco P, Kopka J, Sprenger H, Hincha DK. Molecular signatures associated with increased freezing tolerance due to low temperature memory inArabidopsis. Plant Cell Environ, 2019, 42(3):854-873.
doi: 10.1111/pce.13502 |
| [79] |
Thiebaut F, Hemerly AS, Ferreira PCG. A role for epigenetic regulation in the adaptation and stress responses of Non-model plants. Front Plant Sci, 2019, 10:246.
doi: 10.3389/fpls.2019.00246 pmid: 30881369 |
| [80] |
Song Y, Jia ZF, Hou YK, Ma X, Li LZ, Jin X, An LZ. Roles of DNA methylation in cold priming in Tartary buckwheat. Front Plant Sci, 2020, 11:608540.
doi: 10.3389/fpls.2020.608540 |
| [81] |
Butelli E, Licciardello C, Zhang Y, Liu JJ, Mackay S, Bailey P, Reforgiato-Recupero G, Martin C. Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell, 2012, 24(3):1242-1255.
doi: 10.1105/tpc.111.095232 |
| [82] | Wang JY, Chen XH, Lai ZX. Molecular mechanism and biological functions of plant epigenetic modifications. Chin J Trop Crop, 2020, 41(10):2099-2112. |
| 王静宇, 陈晓慧, 赖钟雄. 植物表观遗传修饰的分子机制及其生物学功能. 热带作物学报, 2020, 41(10):2099-2112. | |
| [83] |
Liu TK, Li Y, Duan WK, Huang FY, Hou XL. Cold acclimation alters DNA methylation patterns and confers tolerance to heat and increases growth rate inBrassica rapa. J Exp Bot, 2017, 68(5):1213-1224.
doi: 10.1093/jxb/erw496 |
| [84] |
Kwon CS, Lee D, Choi G, Chung WI. Histone occupancy-dependent and -independent removal of H3K27 trimethylation at cold-responsive genes in Arabidopsis. Plant J, 2009, 60(1):112-121.
doi: 10.1111/tpj.2009.60.issue-1 |
| [85] |
Saether T, Berge T, Ledsaak M, Matre V, Alm-Kristiansen AH, Dahle O, Aubry F, Gabrielsen OS. The chromatin remodeling factor mi-2α acts as a novel co-activator for human c-Myb. J Biol Chem, 2007, 282(19):13994-14005.
pmid: 17344210 |
| [86] |
Yang R, Hong YC, Ren ZZ, Tang K, Zhang H, Zhu JK, Zhao CZ. A role for PICKLE in the regulation of cold and salt stress tolerance in Arabidopsis. Front Plant Sci, 2019, 10:900.
doi: 10.3389/fpls.2019.00900 |
| [87] |
Shen Y, Lei TT, Cui XY, Liu XY, Zhou SL, Zheng Y, Guérard F, Issakidis-Bourguet E, Zhou DX. Arabidopsis histone deacetylase HDA 15 directly represses plant response to elevated ambient temperature. Plant J, 2019, 100(5):991-1006.
doi: 10.1111/tpj.v100.5 |
| [88] |
Liu HC, Lämke J, Lin SY, Hung MJ, Liu KM, Charng YY, Bäurle I. Distinct heat shock factors and chromatin modifications mediate the organ-autonomous transcriptional memory of heat stress. Plant J, 2018, 95(3):401-413.
doi: 10.1111/tpj.2018.95.issue-3 |
| [89] |
Liu JZ, Feng LL, Gu XT, Deng X, Qiu Q, Li Q, Zhang YY, Wang MY, Deng YW, Wang ET, He YK, Bäurle I, Li JM, Cao XF, He ZH. An H3K27me3 demethylase- HSFA2 regulatory loop orchestrates transgenerational thermomemory in Arabidopsis. Cell Res, 2019, 29(5):379-390.
doi: 10.1038/s41422-019-0145-8 |
| [90] |
Guan QM, Lu XY, Zeng HT, Zhang YY, Zhu JH. Heat stress induction of miR398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. Plant J, 2013, 74(5):840-851.
doi: 10.1111/tpj.2013.74.issue-5 |
| [91] |
Gayacharan, Joel AJ. Epigenetic responses to drought stress in rice (Oryza sativa L.). Physiol Mol Biol Plants, 2013, 19(3):379-387.
doi: 10.1007/s12298-013-0176-4 |
| [92] |
Zheng XG, Chen L, Xia H, Wei HB, Lou QJ, Li MS, Li TM, Luo LJ. Transgenerational epimutations induced by multi-generation drought imposition mediate rice plant’s adaptation to drought condition. Sci Rep, 2017, 7:39843.
doi: 10.1038/srep39843 |
| [93] | Herman JJ, Sultan SE. DNA methylation mediates genetic variation for adaptive transgenerational plasticity. Proc Biol Sci, 2016, 283(1838):20160988. |
| [94] |
Wang GQ, Li HX, Meng S, Yang JC, Ye NH, Zhang JH. Analysis of global methylome and gene expression during carbon reserve mobilization in stems under soil drying. Plant Physiol, 2020, 183(4):1809-1824.
doi: 10.1104/pp.20.00141 |
| [95] |
Benoit M, Drost HG, Catoni M, Gouil Q, Lopez- Gomollon S, Baulcombe D, Paszkowski J. Environmental and epigenetic regulation of rider retrotransposons in tomato. PLoS Genet, 2019, 15(9):e1008370.
doi: 10.1371/journal.pgen.1008370 |
| [96] |
Sukiran NL, Ma JC, Ma H, Su Z. ANAC019 is required for recovery of reproductive development under drought stress in Arabidopsis. Plant Mol Biol, 2019, 99(1-2):161-174.
doi: 10.1007/s11103-018-0810-1 pmid: 30604322 |
| [97] |
Khan IU, Ali A, Khan HA, Baek D, Park J, Lim CJ, Zareen S, Jan M, Lee SY, Pardo JM, Kim WY, Yun DJ. PWR/HDA9/ABI4 complex epigenetically regulates ABA dependent drought stress tolerance in Arabidopsis. Front Plant Sci, 2020, 11:623.
doi: 10.3389/fpls.2020.00623 |
| [98] |
Ding Y, Avramova Z, Fromm M. The Arabidopsis trithorax-like factor ATX1 functions in dehydration stress responses via ABA-dependent and ABA-independent pathways. Plant J, 2011, 66(5):735-744.
doi: 10.1111/j.1365-313X.2011.04534.x |
| [99] |
Liu YT, Zhang A, Yin H, Meng QX, Yu XM, Huang SZ, Wang J, Ahmad R, Liu B, Xu ZY. Trithorax-group proteins ARABIDOPSIS TRITHORAX4 (ATX4) and ATX5 function in abscisic acid and dehydration stress responses. New Phytol, 2018, 217(4):1582-1597.
doi: 10.1111/nph.14933 |
| [100] |
Ding Y, Lapko H, Ndamukong I, Xia YN, Al-Abdallat A, Lalithambika S, Sadder M, Saleh A, Fromm M, Riethoven JJ, Lu GQ, Avramova Z. The Arabidopsis chromatin modifier ATX1, the myotubularin-like AtMTM and the response to drought. Plant Signal Behav, 2009, 4(11):1049-1058.
doi: 10.4161/psb.4.11.10103 |
| [101] |
Ding Y, Fromm M, Avramova Z. Multiple exposures to drought ‘train’ transcriptional responses inArabidopsis. Nat Commun, 2012, 3:740.
doi: 10.1038/ncomms1732 |
| [102] |
Guo TT, Wang DF, Fang JJ, Zhao JF, Yuan SJ, Xiao LT, Li XY. Mutations in the riceOsCHR4 gene, encoding a CHD3 family chromatin remodeler, induce narrow and rolled leaves with increased cuticular wax. Int J Mol Sci, 2019, 20(10):2567.
doi: 10.3390/ijms20102567 |
| [103] |
Li S, Lin YCJ, Wang PY, Zhang BF, Li M, Chen S, Shi R, Tunlaya-Anukit S, Liu XY, Wang ZF, Dai XF, Yu J, Zhou CG, Liu BG, Wang JP, Chiang VL, Li W. The AREB1 transcription factor influences histone acetylation to regulate drought responses and tolerance in Populus trichocarpa. Plant Cell, 2019, 31(3):663-686.
doi: 10.1105/tpc.18.00437 |
| [104] |
Mao HD, Wang HW, Liu SX, Li ZG, Yang XH, Yan JB, Li JS, Tran LSP, Qin F. A transposable element in a NAC gene is associated with drought tolerance in maize seedlings. Nat Commun, 2015, 6:8326.
doi: 10.1038/ncomms9326 |
| [105] | Zhou W, Wang Y, Xie LN. Advances in the establishment and inheritance of plant histone lysine methylation. Biotechnol Bull, 2020, 36(4):117-130. |
| 周伟, 王宇, 解莉楠. 植物组蛋白赖氨酸甲基化建立过程及其遗传性研究进展. 生物技术通报, 2020, 36(4):117-130. | |
| [106] | Liang D, Zhang ZJ, Wu HL, Huang CY, Shuai P, Ye CY, Tang S, Wang YJ, Yang L, Wang J, Yin WL, Xia XL. Single-base-resolution methylomes of Populus trichocarpa reveal the association between DNA methylation and drought stress. BMC Genet, 2014, 15(Suppl 1):S9. |
| [107] |
Han SK, Sang Y, Rodrigues A, BIOL425 F2010, Wu MF, Rodriguez PL, Wagner D. The SWI2/SNF2 chromatin remodeling ATPase BRAHMA represses abscisic acid responses in the absence of the stress stimulus inArabidopsis. Plant Cell, 2012, 24(12):4892-4906.
doi: 10.1105/tpc.112.105114 |
| [108] |
Yang J, Chang Y, Qin YH, Chen DJ, Zhu T, Peng KQ, Wang HJ, Tang N, Li XK, Wang YS, Liu YM, Li XH, Xie WB, Xiong LZ. A lamin-like protein OsNMCP1 regulates drought resistance and root growth through chromatin accessibility modulation by interacting with a chromatin remodeller OsSWI3C in rice. New Phytol, 2020, 227(1):65-83.
doi: 10.1111/nph.16518 pmid: 32129897 |
| [109] |
Baek D, Jiang JF, Chung JS, Wang BS, Chen JP, Xin ZG, Shi HZ. RegulatedAtHKT1 gene expression by a distal enhancer element and DNA methylation in the promoter plays an important role in salt tolerance. Plant Cell Physiol, 2011, 52(1):149-161.
doi: 10.1093/pcp/pcq182 |
| [110] | Dyachenko OV, Zakharchenko NS, Shevchuk TV, Bohnert HJ, Cushman JC, Buryanov YI. Effect of hypermethylation of CCWGG sequences in DNA ofMesembryanthemum crystallinum plants on their adaptation to salt stress. Biochemistry (Mosc), 2006, 71(4):461-465. |
| [111] |
Wang M, Qin LM, Xie C, Li W, Yuan JR, Kong LN, Yu WL, Xia GM, Liu SW. Induced and constitutive DNA methylation in a salinity-tolerant wheat introgression line. Plant Cell Physiol, 2014, 55(7):1354-1365.
doi: 10.1093/pcp/pcu059 |
| [112] | Ogneva ZV, Suprun AR, Dubrovina AS, Kiselev KV. Effect of 5-azacytidine induced DNA demethylation on abiotic stress tolerance in Arabidopsis thaliana. Plant Prot Sci, 2019, 55(2):73-80. |
| [113] |
Bharti P, Mahajan M, Vishwakarma AK, Bhardwaj J, Yadav SK. AtROS1 overexpression provides evidence for epigenetic regulation of genes encoding enzymes of flavonoid biosynthesis and antioxidant pathways during salt stress in transgenic tobacco. J Exp Bot, 2015, 66(19):5959-5969.
doi: 10.1093/jxb/erv304 |
| [114] |
Duan CG, Zhu JK, Cao XF. Retrospective and perspective of plant epigenetics in China. J Genet Genomics, 2018, 45(11):621-638.
doi: 10.1016/j.jgg.2018.09.004 |
| [115] |
Sani E, Herzyk P, Perrella G, Colot V, Amtmann A. Hyperosmotic priming of Arabidopsis seedlings establishes a long-term somatic memory accompanied by specific changes of the epigenome. Genome Biol, 2013, 14(6):R59.
doi: 10.1186/gb-2013-14-6-r59 |
| [116] |
Chen LT, Wu KQ. Role of histone deacetylases HDA6 and HDA19 in ABA and abiotic stress response. Plant Signal Behav, 2010, 5(10):1318-1320.
doi: 10.4161/psb.5.10.13168 |
| [117] |
Veiseth SV, Rahman MA, Yap KL, Fischer A, Egge- Jacobsen W, Reuter G, Zhou MM, Aalen RB, Thorstensen T. The SUVR4 histone lysine methyltransferase binds ubiquitin and converts H3K9me1 to H3K9me3 on transposon chromatin in Arabidopsis. PLoS Genet, 2011, 7(3):e1001325.
doi: 10.1371/journal.pgen.1001325 |
| [118] |
Park HJ, Kim WY, Park HC, Lee SY, Bohnert HJ, Yun DJ. SUMO and SUMOylation in plants. Mol Cells, 2011, 32(4):305-316.
doi: 10.1007/s10059-011-0122-7 pmid: 21912873 |
| [119] | Yuan C, Zhang SW, Niu Y, Tang QL, Wei DY, Wang ZM. Advances in research on the mechanism of DNA methylation in plants. Chin J Biotech, 2020, 36(5):838-848. |
| 袁超, 张少伟, 牛义, 汤青林, 魏大勇, 王志敏. 植物DNA甲基化作用机制的研究进展. 生物工程学报, 2020, 36(5):838-848. | |
| [120] |
Yu A, Lepère G, Jay F, Wang JY, Bapaume L, Wang Y, Abraham AL, Penterman J, Fischer RL, Voinnet O, Navarro L. Dynamics and biological relevance of DNA demethylation in Arabidopsis antibacterial defense. Proc Natl Acad Sci USA, 2013, 110(6):2389-2394.
doi: 10.1073/pnas.1211757110 |
| [121] |
Rodríguez-Negrete EA, Carrillo-Tripp J, Rivera- Bustamante RF. RNA silencing against geminivirus: complementary action of posttranscriptional gene silencing and transcriptional gene silencing in host recovery. J Virol, 2009, 83(3):1332-1340.
doi: 10.1128/JVI.01474-08 |
| [122] |
Li FF, Xu XB, Huang CJ, Gu ZH, Cao LG, Hu T, Ding M, Li ZH, Zhou XP. The AC5 protein encoded by Mungbean yellow mosaic India virus is a pathogenicity determinant that suppresses RNA silencing-based antiviral defenses. New Phytol, 2015, 208(2):555-569.
doi: 10.1111/nph.2015.208.issue-2 |
| [123] |
Luna E, Bruce TJA, Roberts MR, Flors V, Ton J. Next-generation systemic acquired resistance. Plant Physiol, 2012, 158(2):844-853.
doi: 10.1104/pp.111.187468 |
| [124] |
Agorio A, Vera P. ARGONAUTE4 is required for resistance to Pseudomonas syringae in Arabidopsis. Plant Cell, 2007, 19(11):3778-3790.
doi: 10.1105/tpc.107.054494 |
| [125] |
Geng SF, Kong XC, Song GY, Jia ML, Guan JT, Wang F, Qin ZR, Wu L, Lan XJ, Li AL, Mao L. DNA methylation dynamics during the interaction of wheat progenitor Aegilops tauschii with the obligate biotrophic fungus Blumeria graminis f. sp. tritici. New Phytol, 2019, 221(2):1023-1035.
doi: 10.1111/nph.2019.221.issue-2 |
| [126] |
Wada Y, Miyamoto K, Kusano T, Sano H. Association between up-regulation of stress-responsive genes and hypomethylation of genomic DNA in tobacco plants. Mol Genet Genomics, 2004, 271(6):658-666.
pmid: 15148604 |
| [127] | Lee S, Fu FY, Xu SM, Lee SY, Yun DJ, Mengiste T. Global regulation of plant immunity by histone lysine methyl transferases. Plant Cell, 2016, 28(7):1640-1661. |
| [128] |
Berr A, Mccallum EJ, Alioua A, Heintz D, Heitz T, Shen WH. Arabidopsis histone methyltransferase SET DOMAIN GROUP8 mediates induction of the jasmonate/ethylene pathway genes in plant defense response to necrotrophic fungi. Plant Physiol, 2010, 154(3):1403-1414.
doi: 10.1104/pp.110.161497 |
| [129] |
Xia ST, Cheng YT, Huang S, Win J, Soards A, Jinn TL, Jones JDG, Kamoun S, Chen S, Zhang YL, Li X. Regulation of transcription of nucleotide-binding leucine-rich repeat-encoding genes SNC1 and RPP4 via H3K4 trimethylation. Plant Physiol, 2013, 162(3):1694-1705.
doi: 10.1104/pp.113.214551 |
| [130] |
Zhou CH, Zhang L, Duan J, Miki B, Wu KQ. HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. Plant Cell, 2005, 17(4):1196-1204.
doi: 10.1105/tpc.104.028514 |
| [131] |
Alvarez ME, Nota F, Cambiagno DA. Epigenetic control of plant immunity. Mol Plant Pathol, 2010, 11(4):563-576.
doi: 10.1111/j.1364-3703.2010.00621.x pmid: 20618712 |
| [132] | Shi XB, Wang XQ, Che JR, Yang L, Wu JW, Luo BB, Wang MG, Zhong Y, Cheng CZ. Cloning and expression analysis of the CsHAC1 gene in response to Huanglongbing (HLB) infection in sweet orange. Chin J Appl Environ Biol, 2020, 26(2):217-225. |
| 石小保, 王星淇, 车婧如, 杨莉, 伍俊为, 罗彬彬, 王梦鸽, 钟云, 程春振. 甜橙CsHAC1基因克隆及其在响应柑橘黄龙病侵染过程中的表达. 应用与环境生物学报, 2020, 26(2):217-225. | |
| [133] |
Tian L, Chen ZJ. Blocking histone deacetylation in Arabidopsis induces pleiotropic effects on plant gene regulation and development. Proc Natl Acad Sci USA, 2001, 98(1):200-205.
doi: 10.1073/pnas.98.1.200 |
| [134] |
Pramanik D, Shelake RM, Kim MJ, Kim JY. CRISPR- mediated engineering across the central dogma in plant biology for basic research and crop improvement. Mol Plant, 2021, 14(1):127-150.
doi: 10.1016/j.molp.2020.11.002 |
| [135] |
Wu K, Tian L, Malik K, Brown D, Miki B. Functional analysis of HD2 histone deacetylase homologues in Arabidopsis thaliana. Plant J, 2000, 22(1):19-27.
pmid: 10792817 |
| [136] |
Deng YW, Zhai KR, Xie Z, Yang DY, Zhu XD, Liu JZ, Wang X, Qin P, Yang YZ, Zhang GM, Li Q, Zhang JF, Wu SQ, Milazzo J, Mao BZ, Wang ET, Xie HA, Tharreau D, He ZH. Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science, 2017, 355(6328):962-965.
doi: 10.1126/science.aai8898 |
| [137] |
Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JDG. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science, 2006, 312(5772):436-439.
pmid: 16627744 |
| [138] |
Katiyar-Agarwal S, Morgan R, Dahlbeck D, Borsani O, Villegas A, Zhu JK, Staskawicz BJ, Jin HL. A pathogen- inducible endogenous siRNA in plant immunity. Proc Natl Acad Sci USA, 2006, 103(47):18002-18007.
doi: 10.1073/pnas.0608258103 |
| [139] |
Andika IB, Sun LY, Xiang R, Li JM, Chen JP. Root-specific role for Nicotiana benthamiana RDR6 in the inhibition of Chinese wheat mosaic virus accumulation at higher temperatures. Mol Plant Microbe Interact, 2013, 26(10):1165-1175.
doi: 10.1094/MPMI-05-13-0137-R |
| [140] |
Hepburn PA, Margison GP, Tisdale MJ. Enzymatic methylation of cytosine in DNA is prevented by adjacent O6-methylguanine residues. J Biol Chem, 1991, 266(13):7985-7987.
pmid: 2022628 |
| [141] | Wang T, Chen ML, Liu L, Ning CL, Cai BH, Zhang Z, Qiao YS. Changes in genome and gene expression during plant polyploidization. Chin Bull Bot, 2015, 50(4):504-515. |
| 王涛, 陈孟龙, 刘玲, 宁传丽, 蔡斌华, 章镇, 乔玉山. 植物多倍体化中基因组和基因表达的变化. 植物学报, 2015, 50(4):504-515. | |
| [142] |
Lauss K, Wardenaar R, Oka R, van Hulten MHA, Guryev V, Keurentjes JJB, Stam M, Johannes F. Parental DNA methylation states are associated with heterosis in epigenetic hybrids. Plant Physiol, 2018, 176(2):1627-1645.
doi: 10.1104/pp.17.01054 |
| [143] |
Zhang MS, Yan HY, Zhao N, Lin XY, Pang JS, Xu KZ, Liu LX, Liu B. Endosperm-specific hypomethylation, and meiotic inheritance and variation of DNA methylation level and pattern in sorghum ( Sorghum bicolor L.) inter-strain hybrids. Theor Appl Genet, 2007, 115(2):195-207.
pmid: 17486309 |
| [144] | Zhao Y, Yu S, Xing C, Fan S, Song M. DNA methylation in cotton hybrids and their parents. Mol Biol (Mosk), 2008, 42(2):195-205. |
| [145] | Peng H, Jiang GH, Zhang J, Zhang WX, Zhai WX. DNA methylation polymorphism and stability in Chinese indica hybrid rice. Sci China Life Sci, 2014, 44(1):45-53. |
| 彭海, 江光怀, 张静, 章伟雄, 翟文学. 中国杂交籼稻DNA甲基化多样性与遗传稳定性. 中国科学: 生命科学, 2014, 44(1):45-53. | |
| [146] |
Seifert F, Thiemann A, Grant-Downton R, Edelmann S, Rybka D, Schrag TA, Frisch M, Dickinson HG, Melchinger AE, Scholten S. Parental expression variation of small RNAs is negatively correlated with grain yield heterosis in a maize breeding population. Front Plant Sci, 2018, 9:13.
doi: 10.3389/fpls.2018.00013 pmid: 29441076 |
| [147] |
Chen ZJ. Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant Sci, 2010, 15(2):57-71.
doi: 10.1016/j.tplants.2009.12.003 |
| [148] |
Groszmann M, Greaves IK, Albertyn ZI, Scofield GN, Peacock WJ, Dennis ES. Changes in 24-nt siRNA levels in Arabidopsis hybrids suggest an epigenetic contribution to hybrid vigor. Proc Natl Acad Sci USA, 2011, 108(6):2617-2622.
doi: 10.1073/pnas.1019217108 |
| [149] |
Barber WT, Zhang W, Win H, Varala KK, Dorweiler JE, Hudson ME, Moose SP. Repeat associated small RNAs vary among parents and following hybridization in maize. Proc Natl Acad Sci USA, 2012, 109(26):10444-10449.
doi: 10.1073/pnas.1202073109 |
| [150] |
Ng DWK, Lu J, Chen ZJ. Big roles for small RNAs in polyploidy, hybrid vigor, and hybrid incompatibility. Curr Opin Plant Biol, 2012, 15(2):154-161.
doi: 10.1016/j.pbi.2012.01.007 |
| [151] |
He GM, Zhu XP, Elling AA, Chen LB, Wang XF, Guo L, Liang MZ, He H, Zhang HY, Chen FF, Qi YJ, Chen RS, Deng XW. Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant cell, 2010, 22(1):17-33.
doi: 10.1105/tpc.109.072041 |
| [152] |
Greaves IK, Groszmann M, Ying H, Taylor JM, Peacock WJ, Dennis ES. Trans chromosomal methylation in Arabidopsis hybrids. Proc Natl Acad Sci USA, 2012, 109(9):3570-3575.
doi: 10.1073/pnas.1201043109 |
| [153] | Cui HH, Xiang C, Shi YR, Wang WS, Gao YM. The research progress of epigenetic insights into bases of heterosis. J Plant Genet Resour, 2015, 16(5):933-939. |
| 崔会会, 项超, 石英尧, 王文生, 高用明. 杂种优势形成的表观遗传学研究进展. 植物遗传资源学报, 2015, 16(5):933-939. | |
| [154] |
Ni ZF, Kim ED, Ha M, Lackey E, Liu JX, Zhang YR, Sun QX, Chen ZJ. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature, 2009, 457(7227):327-331.
doi: 10.1038/nature07523 |
| [155] |
Wang JL, Tian L, Lee HS, Chen ZJ. Nonadditive regulation of FRI and FLC loci mediates flowering-time variation in Arabidopsis allopolyploids. Genetics, 2006, 173(2):965-974.
doi: 10.1534/genetics.106.056580 |
| [156] |
Moghaddam AMB, Roudier F, Seifert M, Bérard C, Magniette MLM, Ashtiyani RK, Houben A, Colot V, Mette MF. Additive inheritance of histone modifications in Arabidopsis thaliana intra-specific hybrids. Plant J, 2011, 67(4):691-700.
doi: 10.1111/tpj.2011.67.issue-4 |
| [157] |
Liu JZ, He ZH. Epigenetic regulation of heat stress response in plants. Chin Sci Bull, 2014, 59(8):631-639.
doi: 10.1360/972013-1103 |
|
刘军钟, 何祖华. 植物响应高温胁迫的表观遗传调控. 科学通报, 2014, 59(8):631-639.
doi: 10.1360/972013-1103 |
|
| [158] |
Lämke J, Bäurle I. Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biol, 2017, 18(1):124.
doi: 10.1186/s13059-017-1263-6 pmid: 28655328 |
| [159] |
Mathieu O, Reinders J, Caikovski M, Smathajitt C, Paszkowski J. Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell, 2007, 130(5):851-862.
doi: 10.1016/j.cell.2007.07.007 |
| [160] |
van Hulten M, Pelser M, van Loon LC, Pieterse CMJ, Ton J. Costs and benefits of priming for defense in Arabidopsis. Proc Natl Acad Sci USA, 2006, 103(14):5602-5607.
doi: 10.1073/pnas.0510213103 |
| [161] |
Springer NM, Schmitz RJ. Exploiting induced and natural epigenetic variation for crop improvement. Nat Rev Genet, 2017, 18(9):563-575.
doi: 10.1038/nrg.2017.45 pmid: 28669983 |
| [162] |
Mcclintock B. The significance of responses of the genome to challenge. Science, 1984, 226(4676):792-801.
doi: 10.1126/science.15739260 |
| [163] |
Lin T, Zhu GT, Zhang JH, Xu XY, Yu QH, Zheng Z, Zhang ZH, Lun YY, Li S, Wang XX, Huang ZJ, Li JM, Zhang CZ, Wang TT, Zhang YY, Wang AX, Zhang YC, Lin K, Li CY, Xiong GS, Xue YB, Mazzucato A, Causse M, Fei ZJ, Giovannoni JJ, Chetelat RT, Zamir D, Städler T, Li JF, Ye ZB, Du YC, Huang SW. Genomic analyses provide insights into the history of tomato breeding. Nat Genet, 2014, 46(11):1220-1226.
doi: 10.1038/ng.3117 |
| [164] |
Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature, 2012, 485(7400):635-641.
doi: 10.1038/nature11119 |
| [165] |
Chen WW, Kong JH, Qin C, Yu S, Tan JJ, Chen YR, Wu CQ, Wang H, Shi Y, Li CY, Li B, Zhang PC, Wang Y, Lai TF, Yu ZM, Zhang X, Shi NN, Wang HZ, Osman T, Liu YL, Manning K, Jackson S, Rolin D, Zhong SL, Seymour GB, Gallusci P, Hong YG. Requirement of CHROMOMETHYLASE3 for somatic inheritance of the spontaneous tomato epimutation Colourless non-ripening. Sci Rep, 2015, 5:9192.
doi: 10.1038/srep09192 |
| [166] |
Morrell PL, Buckler ES, Ross-Ibarra J. Crop genomics: advances and applications. Nat Rev Genet, 2011, 13(2):85-96.
doi: 10.1038/nrg3097 pmid: 22207165 |
| [167] |
Martin A, Troadec C, Boualem A, Rajab M, Fernandez R, Morin H, Pitrat M, Dogimont C, Bendahmane A. A transposon-induced epigenetic change leads to sex determination in melon. Nature, 2009, 461(7267):1135-1138.
doi: 10.1038/nature08498 |
| [168] |
Miura K, Agetsuma M, Kitano H, Yoshimura A, Matsuoka M, Jacobsen SE, Ashikari M. A metastable DWARF1 epigenetic mutant affecting plant stature in rice. Proc Natl Acad Sci USA, 2009, 106(27):11218-11223.
doi: 10.1073/pnas.0901942106 |
| [1] | 许梦萱, 周明. 植物RNA聚合酶IV调控DNA甲基化和发育的研究进展[J]. 遗传, 2022, 44(7): 567-580. |
| [2] | 曲卉, 柳毅, 陈雅文, 汪晖. 环境因素所致印迹基因改变与子代器官发育[J]. 遗传, 2022, 44(2): 107-116. |
| [3] | 张杨景晖, 常沛瑶, 杨紫淑, 薛宇航, 李雪奇, 张旸. 表观遗传修饰影响花青苷合成研究进展[J]. 遗传, 2022, 44(12): 1117-1127. |
| [4] | 张子寅, 周燕萍, 孟卓贤. CUT&Tag技术在代谢组织细胞的实验操作[J]. 遗传, 2022, 44(10): 958-966. |
| [5] | 赵清雯, 潘东宁. 表观遗传修饰对脂肪组织产热的调控进展[J]. 遗传, 2022, 44(10): 867-880. |
| [6] | 王天一, 王应祥, 尤辰江. 植物PHD结构域蛋白的结构与功能特性[J]. 遗传, 2021, 43(4): 323-339. |
| [7] | 张向前, 李楠, 解新明. 表观遗传学综合性实验设计与探讨[J]. 遗传, 2021, 43(12): 1179-1187. |
| [8] | 王芯悦, 李亮, 段秋慧, 李大力, 陈金联. Uhrf1对肠上皮发育的影响[J]. 遗传, 2021, 43(1): 84-93. |
| [9] | 吴杰, 全建平, 叶勇, 吴珍芳, 杨杰, 杨明, 郑恩琴. 染色质转座酶可及性测序研究进展[J]. 遗传, 2020, 42(4): 333-346. |
| [10] | 崔亨贞, 孙蜜烛, 王润芝, 李辰雨, 黄予暄, 黄秋菊, 乔晓孟. 内侧前额叶皮质DNA甲基化调控大鼠酒精相关行为[J]. 遗传, 2020, 42(1): 112-125. |
| [11] | 王昕源, 张雨, 杨楠, 程禾, 孙玉洁. DNMT3a通过提升基因内部甲基化介导紫杉醇诱导的LINE-1异常表达[J]. 遗传, 2020, 42(1): 100-111. |
| [12] | 张競文,续倩,李国亮. 癌症发生发展中的表观遗传学研究[J]. 遗传, 2019, 41(7): 567-581. |
| [13] | 黄鑫,陈永强,徐国良,彭淑红. 脂肪组织DNA甲基化与糖尿病和肥胖的发生发展[J]. 遗传, 2019, 41(2): 98-110. |
| [14] | 杨旭琼, 吴珍芳, 李紫聪. 哺乳动物体细胞核移植表观遗传重编程研究进展[J]. 遗传, 2019, 41(12): 1099-1109. |
| [15] | 潘云枫, 王演怡, 陈静雯, 范怡梅. 线粒体代谢介导的表观遗传改变与衰老研究[J]. 遗传, 2019, 41(10): 893-904. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||

www.chinagene.cn
备案号:京ICP备09063187号-4
总访问:,今日访问:,当前在线:
