遗传 ›› 2025, Vol. 47 ›› Issue (8): 944-957.doi: 10.16288/j.yczz.25-030
• 综述 • 上一篇
收稿日期:2025-01-26
修回日期:2025-04-30
出版日期:2025-08-20
发布日期:2025-05-07
通讯作者:
任国栋,研究员,博士生导师,研究方向:植物遗传与RNA生物学。E-mail: gdren@fudan.edu.cn作者简介:郭梦玮,博士研究生,专业方向:生物化学与生物物理学。E-mail: 20110700093@fudan.edu.cn郭梦玮和樊友宏并列第一作者。
基金资助:
Mengwei Guo( ), Youhong Fan(
), Youhong Fan( ), Guodong Ren(
), Guodong Ren( )
)
Received:2025-01-26
Revised:2025-04-30
Published:2025-08-20
Online:2025-05-07
Supported by:摘要:
miRNA (microRNA)是一类由内源基因编码的、长度在20~24个核苷酸的小分子非编码RNA。miRNA主要在转录后水平调控基因表达,进而影响动植物生殖、发育和环境应答等各种生物学过程。miRNA在不同组织和细胞内的分布、稳态维持和动态调节受到多个层次调控,包括转录、加工生成、稳定性调节以及靶向降解等。关于miRNA生物合成(包括转录和加工)的生化途径已经建立,对其调控的分子机制也有了较为深入的认识。本文系统综述了植物miRNA生成后稳定性调节、周转和靶向降解的相关研究进展,并结合动物中的机制进行比较和讨论,旨在为深入阐明控制细胞内miRNA丰度的分子机制提供理论框架。
郭梦玮, 樊友宏, 任国栋. 植物miRNA稳定性与降解的分子基础[J]. 遗传, 2025, 47(8): 944-957.
Mengwei Guo, Youhong Fan, Guodong Ren. Molecular basis of microRNA stability and degradation in plants[J]. Hereditas(Beijing), 2025, 47(8): 944-957.
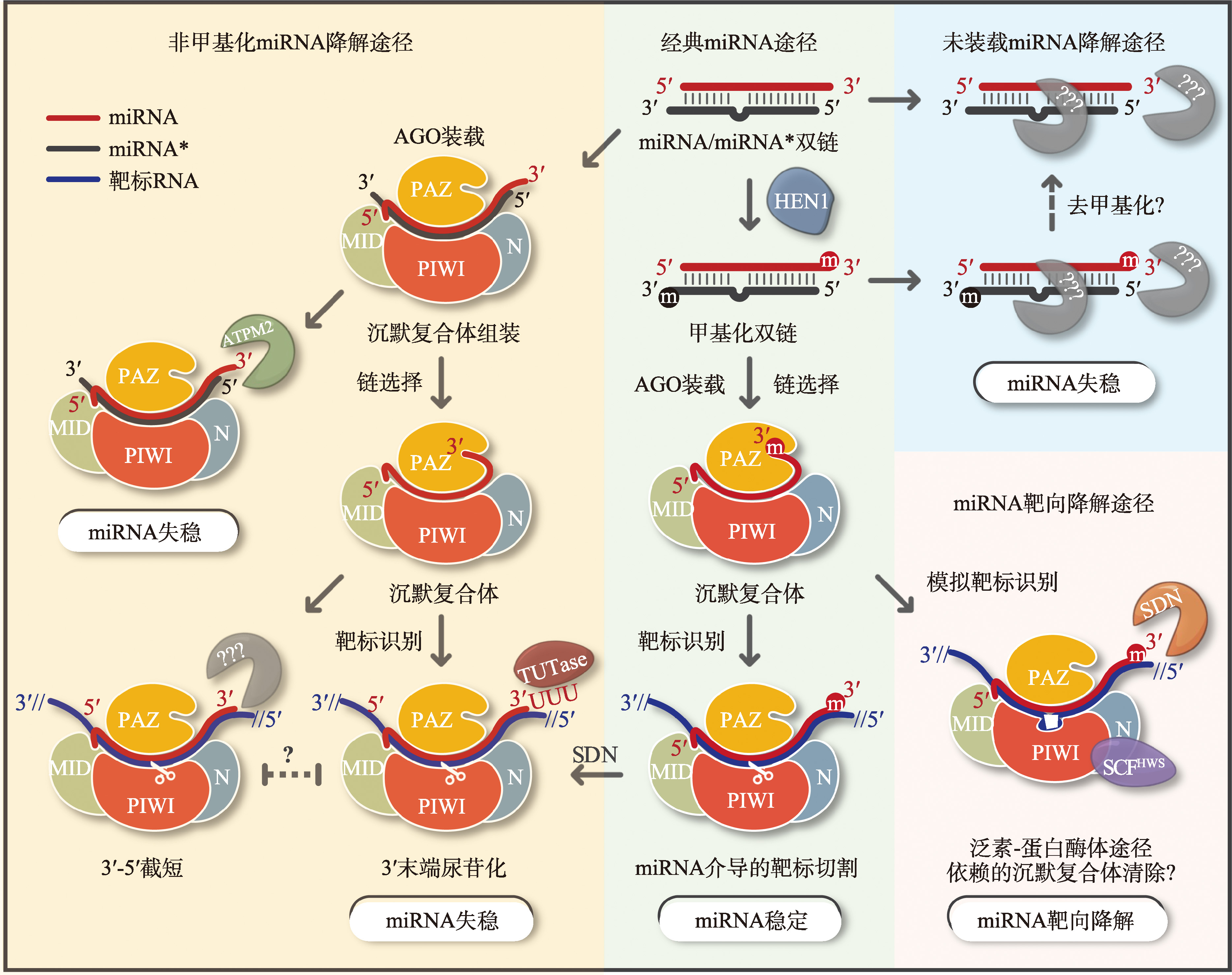
图1
植物miRNA稳定性调节与降解代谢的工作模式图 植物中新生miRNA/miRNA*经HEN1介导的2′-O-甲基化修饰后,miRNA链被选择性地组装到AGO蛋白中形成沉默复合体(RISC),RISC中的miRNA通过序列配对识别并抑制靶基因(此处用靶标切割表示,也存在翻译抑制)的表达。末端甲基化修饰及AGO的结合对于miRNA的稳定性至关重要。未组装的miRNA/miRNA*通过未知的核酸酶降解。3′-5′核酸外切酶ATRM2与AGO1相互作用,参与部分未甲基化miRNA/miRNA*的降解,推测该过程可能发生在RISC装载的初始阶段。和动物中的种子区配对不同,植物内源靶标与miRNA通常高度配对,这使得miRNA的3′末端从AGO蛋白的PAZ结构域中脱离出来。在这种情况下,末端尿苷转移酶TUTase (如拟南芥中的HESO1和URT1、衣藻中的MUT68)以及未鉴定到的3′-5′核酸外切酶相互竞争并攻击miRNA 3′末端,引起miRNA 3′末端的加尾与截短。在这个过程中,多尿苷化加尾促进miRNA的降解,而截短的生物学意义仍不清楚。甲基化可以阻止加尾和截短,维持miRNA 3′末端的完整性。SDN家族3′-5′核酸外切酶可能通过切除含甲基化修饰的miRNA,使其进入加尾、截短依赖的miRNA失稳降解。模拟靶标是一类在miRNA-靶标配对中间区域有凸出泡的特殊靶标,并使得RISC无法对模拟靶标进行切割。相反,该凸出泡可能通过未知的机制引发RISC构象或翻译后修饰的改变,进而招募F-box家族蛋白HWS,引起RISC复合物的降解。SDN1/2也被报道参与该过程,但具体的分子机制目前仍不清楚。RISC: RNA诱导沉默复合体(RNA-induced silencing complex);TUTase: 末端尿苷转移酶(terminal uridylyl transferase);SCF,SKP1-CUL1-F-box泛素连接酶;UPS: 泛素-蛋白酶体系统(ubiquitin-proteasome system)。"
| [1] | Tian W, Chen T, Liu QY, Zhang BS, Guo HS, Zhao JH. Advances in the mechanisms and applications of RNA silencing in crop protection. Hereditas(Beijing), 2024, 46(4): 266-278. |
| 田文, 谌婷, 刘清艳, 张博森, 郭惠珊, 赵建华. 植物RNA沉默抗病机制与应用研究进展. 遗传, 2024, 46(4): 266-278. | |
| [2] |
Wang JL, Mei J, Ren GD. Plant microRNAs: biogenesis, homeostasis, and degradation. Front Plant Sci, 2019, 10: 360.
pmid: 30972093 |
| [3] |
Axtell MJ. Classification and comparison of small RNAs from plants. Annu Rev Plant Biol, 2013, 64: 137-159.
pmid: 23330790 |
| [4] |
Kim VN, Han JJ, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol, 2009, 10(2): 126-139.
pmid: 19165215 |
| [5] |
Cuperus JT, Fahlgren N, Carrington JC. Evolution and functional diversification of MIRNA genes. Plant Cell, 2011, 23(2): 431-442.
pmid: 21317375 |
| [6] |
Rogers K, Chen XM. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell, 2013, 25(7): 2383-2399.
pmid: 23881412 |
| [7] |
Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Gene Dev, 2006, 20(24): 3407-3425.
pmid: 17182867 |
| [8] |
Yang ZY, Ebright YW, Yu B, Chen XM. HEN1 recognizes 21-24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Res, 2006, 34(2): 667-675.
pmid: 16449203 |
| [9] |
Yu B, Yang ZY, Li JJ, Minakhina S, Yang MC, Padgett RW, Steward R, Chen XM. Methylation as a crucial step in plant microRNA biogenesis. Science, 2005, 307(5711): 932-935.
pmid: 15705854 |
| [10] |
Iki T, Yoshikawa M, Meshi T, Ishikawa M. Cyclophilin 40 facilitates HSP90-mediated RISC assembly in plants. EMBO J, 2012, 31(2): 267-278.
pmid: 22045333 |
| [11] |
Bologna NG, Iselin R, Abriata LA, Sarazin A, Pumplin N, Jay F, Grentzinger T, Dal Peraro M, Voinnet O. Nucleo-cytosolic shuttling of ARGONAUTE1 prompts a revised model of the plant microRNA pathway. Mol Cell, 2018, 69(4): 709-719.e5.
pmid: 29398448 |
| [12] |
Bartel DP. Metazoan microRNAs. Cell, 2018, 173(1): 20-51.
pmid: 29570994 |
| [13] |
Axtell MJ, Bowman JL. Evolution of plant microRNAs and their targets. Trends Plant Sci, 2008, 13(7): 343-349.
pmid: 18502167 |
| [14] |
Creasey KM, Zhai JX, Borges F, Van Ex F, Regulski M, Meyers BC, Martienssen RA. MiRNAs trigger widespread epigenetically activated siRNAs from transposons in Arabidopsis. Nature, 2014, 508(7496): 411-415.
pmid: 24670663 |
| [15] |
Xue Y, Cao XF, Chen XS, Deng X, Deng XW, Ding Y, Dong AW, Duan CG, Fang XF, Gong L, Gong ZZ, Gu XF, He CS, He H, He SB, He XJ, He Y, He YH, Jia GF, Jiang DH, Jiang JJ, Lai JS, Lang ZB, Li CL, Li Q, Li XW, Liu B, Liu B, Luo X, Qi YJ, Qian WQ, Ren GD, Song QX, Song XW, Tian ZX, Wang JW, Wang Y, Wu L, Wu Z, Xia R, Xiao J, Xu L, Xu ZY, Yan WH, Yang HC, Zhai JX, Zhang YJ, Zhao YS, Zhong XH, Zhou DX, Zhou M, Zhou Y, Zhu B, Zhu JK, Liu QK. Epigenetics in the modern era of crop improvements. Sci China Life Sci, 2025, doi: 10.1007/s11427-024-2784-3.
pmid: 39808224 |
| [16] |
Song XW, Li Y, Cao XF, Qi YJ. MicroRNAs and their regulatory roles in plant-environment interactions. Annu Rev Plant Biol, 2019, 70: 489-525.
pmid: 30848930 |
| [17] | Xu J, Hou N, Han N, Bian HW, Zhu MY. The regulatory roles of small RNAs in phytohormone signaling pathways. Hereditas(Beijing), 2016, 38(5): 418-426. |
| 许佳, 侯宁, 韩凝, 边红武, 朱睦元. 小分子RNA在植物激素信号通路中的调控功能. 遗传, 2016, 38(5): 418-426. | |
| [18] |
Park W, Li JJ, Song RT, Messing J, Chen XM. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol, 2002, 12(17): 1484-1495.
pmid: 12225663 |
| [19] |
Li SJ, Castillo-González C, Yu B, Zhang XR. The functions of plant small RNAs in development and in stress responses. Plant J, 2017, 90(4): 654-670.
pmid: 27943457 |
| [20] |
Chen XM, Rechavi O. Plant and animal small RNA communications between cells and organisms. Nat Rev Mol Cell Biol, 2022, 23(3): 185-203.
pmid: 34707241 |
| [21] |
Xu Y, Chen XM. MicroRNA biogenesis and stabilization in plants. Fundam Res, 2023, 3(5): 707-717.
pmid: 38933298 |
| [22] |
Fang XF, Qi YJ. RNAi in plants: An Argonaute-centered view. Plant Cell, 2016, 28(2): 272-285.
pmid: 26869699 |
| [23] |
Meister G. Argonaute proteins: functional insights and emerging roles. Nat Rev Genet, 2013, 14(7): 447-459.
pmid: 23732335 |
| [24] |
Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet, 2011, 12(1): 19-31.
pmid: 21116305 |
| [25] |
Mi SJ, Cai T, Hu YG, Chen YM, Hodges E, Ni FR, Wu L, Li S, Zhou HY, Long CZ, Chen S, Hannon GJ, Qi YJ. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5' terminal nucleotide. Cell, 2008, 133(1): 116-127.
pmid: 18342361 |
| [26] |
Frank F, Sonenberg N, Nagar B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature, 2010, 465(7299): 818-822.
pmid: 20505670 |
| [27] |
Schwarz DS, Hutvágner G, Du TT, Xu ZS, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell, 2003, 115(2): 199-208.
pmid: 14567917 |
| [28] |
Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell, 2003, 115(2): 209-216.
pmid: 14567918 |
| [29] |
Vaucheret H, Vazquez F, CrétéP, Bartel DP. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Gene Dev, 2004, 18(10): 1187-1197.
pmid: 15131082 |
| [30] |
Giudicatti AJ, Tomassi AH, Manavella PA, Arce AL. Extensive analysis of miRNA trimming and tailing indicates that AGO1 has a complex role in miRNA turnover. Plants (Basel), 2021, 10(2): 267.
pmid: 33573197 |
| [31] |
Li SB, Le B, Ma X, Li SF, You CJ, Yu Y, Zhang BL, Liu L, Gao L, Shi T, Zhao YH, Mo BX, Cao XF, Chen XM. Biogenesis of phased siRNAs on membrane-bound polysomes in Arabidopsis. eLife, 2016, 5: e22750.
pmid: 27938667 |
| [32] |
Zhai JX, Zhao YY, Simon SA, Huang S, Petsch K, Arikit S, Pillay M, Ji LJ, Xie M, Cao XF, Yu B, Timmermans M, Yang B, Chen XM, Meyers BC. Plant microRNAs display differential 3′ truncation and tailing modifications that are ARGONAUTE1 dependent and conserved across species. Plant Cell, 2013, 25(7): 2417-2428.
pmid: 23839787 |
| [33] |
Montgomery TA, Howell MD, Cuperus JT, Li DW, Hansen JE, Alexander AL, Chapman EJ, Fahlgren N, Allen E, Carrington JC. Specificity of ARGONAUTE7- miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell, 2008, 133(1): 128-141.
pmid: 18342362 |
| [34] |
Li J, Wang Z, Hu YG, Cao Y, Ma LG. Polycomb group proteins RING1A and RING1B regulate the vegetative phase transition in Arabidopsis. Front Plant Sci, 2017, 8: 867.
pmid: 28596781 |
| [35] |
Yifhar T, Pekker I, Peled D, Friedlander G, Pistunov A, Sabban M, Wachsman G, Alvarez JP, Amsellem Z, Eshed Y. Failure of the tomato trans-acting short interfering RNA program to regulate AUXIN RESPONSE FACTOR3 and ARF4 underlies the wiry leaf syndrome. Plant Cell, 2012, 24(9): 3575-3589.
pmid: 23001036 |
| [36] |
Ji LJ, Liu XG, Yan J, Wang WM, Yumul RE, Kim YJ, Dinh TT, Liu J, Cui X, Zheng BL, Agarwal M, Liu CY, Cao XF, Tang GL, Chen XM. ARGONAUTE10 and ARGONAUTE1 regulate the termination of floral stem cells through two microRNAs in Arabidopsis. PLoS Genet, 2011, 7(3): e1001358.
pmid: 21483759 |
| [37] |
Zhu HL, Hu FQ, Wang RH, Zhou X, Sze SH, Liou LW, Barefoot A, Dickman M, Zhang XR. Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell, 2011, 145(2): 242-256.
pmid: 21496644 |
| [38] |
Yu Y, Ji LJ, Le BH, Zhai JX, Chen JY, Luscher E, Gao L, Liu CY, Cao XF, Mo BX, Ma JB, Meyers BC, Chen XM. ARGONAUTE10 promotes the degradation of miR165/6 through the SDN1 and SDN2 exonucleases in Arabidopsis. PLoS Biol, 2017, 15(2): e2001272.
pmid: 28231321 |
| [39] | Baccarini A, Chauhan H, Gardner TJ, Jayaprakash AD, Sachidanandam R, Brown BD. Kinetic analysis reveals the fate of a microRNA following target regulation in mammalian cells. Curr Biol, 2011, 21(5): 369-376. [DOI] |
| [40] |
Derrien B, Baumberger N, Schepetilnikov M, Viotti C, De Cillia J, Ziegler-Graff V, Isono E, Schumacher K, Genschik P. Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc Natl Acad Sci USA, 2012, 109(39): 15942-15946.
pmid: 23019378 |
| [41] |
Michaeli S, Clavel M, Lechner E, Viotti C, Wu J, Dubois M, Hacquard T, Derrien B, Izquierdo E, Lecorbeiller M, Bouteiller N, De Cilia J, Ziegler-Graff V, Vaucheret H, Galili G, Genschik P. The viral F-box protein P0 induces an ER-derived autophagy degradation pathway for the clearance of membrane-bound AGO1. Proc Natl Acad Sci USA, 2019, 116(45): 22872-22883.
pmid: 31628252 |
| [42] |
Earley K, Smith M, Weber R, Gregory B, Poethig R. An endogenous F-box protein regulates ARGONAUTE1 in Arabidopsis thaliana. Silence, 2010, 1(1): 15.
pmid: 20624295 |
| [43] |
Hacquard T, Clavel M, Baldrich P, Lechner E, Pérez-Salamó I, Schepetilnikov M, Derrien B, Dubois M, Hammann P, Kuhn L, Brun D, Bouteiller N, Baumberger N, Vaucheret H, Meyers BC, Genschik P. The Arabidopsis F-box protein FBW2 targets AGO1 for degradation to prevent spurious loading of illegitimate small RNA. Cell Rep, 2022, 39(2): 110671.
pmid: 35417704 |
| [44] |
Ré DA, Cambiagno DA, Arce AL, Tomassi AH, Giustozzi M, Casati P, Ariel FD, Manavella PA. CURLY LEAF regulates microRNA activity by controlling ARGONAUTE 1 degradation in plants. Mol Plant, 2020, 13(1): 72-87.
pmid: 31606467 |
| [45] |
Chen XM, Liu J, Cheng YL, Jia DX. HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development, 2002, 129(5): 1085-1094.
pmid: 11874905 |
| [46] |
Huang Y, Ji LJ, Huang QC, Vassylyev DG, Chen XM, Ma JB. Structural insights into mechanisms of the small RNA methyltransferase HEN1. Nature, 2009, 461(7265): 823-827.
pmid: 19812675 |
| [47] |
Abe M, Yoshikawa T, Nosaka M, Sakakibara H, Sato Y, Nagato Y, Itoh J. WAVY LEAF1, an ortholog of Arabidopsis HEN1, regulates shoot development by maintaining MicroRNA and trans-acting small interfering RNA accumulation in rice. Plant Physiol, 2010, 154(3): 1335-1346.
pmid: 20805329 |
| [48] |
Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol, 2013, 14(8): 475-488.
pmid: 23800994 |
| [49] |
Ren GD, Chen XM, Yu B. Small RNAs meet their targets: when methylation defends miRNAs from uridylation. RNA Biol, 2014, 11(9): 1099-1104.
pmid: 25483033 |
| [50] |
Li JJ, Yang ZY, Yu B, Liu J, Chen XM. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol, 2005, 15(16): 1501-1507.
pmid: 16111943 |
| [51] |
Ren GD, Chen XM, Yu B. Uridylation of miRNAs by hen1 suppressor1 in Arabidopsis. Curr Biol, 2012, 22(8): 695-700.
pmid: 22464191 |
| [52] |
Tu B, Liu L, Xu C, Zhai JX, Li SB, Lopez MA, Zhao YY, Yu Y, Ramachandran V, Ren GD, Yu B, Li SG, Meyers BC, Mo BX, Chen XM. Distinct and cooperative activities of HESO1 and URT1 nucleotidyl transferases in microRNA turnover in Arabidopsis. PLoS Genet, 2015, 11(4): e1005119.
pmid: 25928405 |
| [53] |
Zhao YY, Yu Y, Zhai JX, Ramachandran V, Dinh TT, Meyers BC, Mo BX, Chen XM. The Arabidopsis nucleotidyl transferase HESO1 uridylates unmethylated small RNAs to trigger their degradation. Curr Biol, 2012, 22(8): 689-694.
pmid: 22464194 |
| [54] |
Wang XY, Zhang SX, Dou YC, Zhang C, Chen XM, Yu B, Ren GD. Synergistic and independent actions of multiple terminal nucleotidyl transferases in the 3' tailing of small RNAs in Arabidopsis. PLoS Genet, 2015, 11(4): e1005091.
pmid: 25928341 |
| [55] |
Hu Q, Yang HR, Li MW, Zhu LR, Lv MQ, Li FD, Zhang ZY, Ren GD, Gong QG. Molecular mechanism underlying the di-uridylation activity of Arabidopsis TUTase URT1. Nucleic Acids Res, 2022, 50(18): 10614-10625.
pmid: 36177876 |
| [56] |
Ren GD, Xie M, Zhang SX, Vinovskis C, Chen XM, Yu B. Methylation protects microRNAs from an AGO1- associated activity that uridylates 5' RNA fragments generated by AGO1 cleavage. Proc Natl Acad Sci USA, 2014, 111(17): 6365-6370.
pmid: 24733911 |
| [57] |
Chen SS, Cai YC, Yang HR, Zhang B, Li N, Ren GD. PBOX-sRNA-seq uncovers novel features of miRNA modification and identifies selected 5′-tRNA fragments bearing 2′-O-modification. Nucleic Acids Res, 2024, 52(14): e65.
pmid: 38908023 |
| [58] |
Song JB, Wang XY, Song B, Gao L, Mo XW, Yue LM, Yang HQ, Lu JY, Ren GD, Mo BX, Chen XM. Prevalent cytidylation and uridylation of precursor miRNAs in Arabidopsis. Nat Plants, 2019, 5(12): 1260-1272.
pmid: 31792392 |
| [59] |
Kong WW, Dong XX, Ren YB, Wang Y, Xu XT, Mo BX, Yu Y, Wang XY. NTP4 modulates miRNA accumulation via asymmetric modification of miRNA/miRNA* duplex. Sci China Life Sci, 2021, 64(5): 832-835.
pmid: 32915408 |
| [60] |
Ibrahim F, Rymarquis LA, Kim EJ, Becker J, Balassa E, Green PJ, Cerutti H. Uridylation of mature miRNAs and siRNAs by the MUT68 nucleotidyltransferase promotes their degradation in Chlamydomonas. Proc Natl Acad Sci USA, 2010, 107(8): 3906-3911.
pmid: 20142471 |
| [61] |
Lu SF, Sun YH, Chiang VL. Adenylation of plant miRNAs. Nucleic Acids Res, 2009, 37(6): 1878-1885.
pmid: 19188256 |
| [62] |
Han J, Mendell JT. MicroRNA turnover: a tale of tailing, trimming, and targets. Trends Biochem Sci, 2023, 48(1): 26-39.
pmid: 35811249 |
| [63] |
Yu S, Kim VN. A tale of non-canonical tails: gene regulation by post-transcriptional RNA tailing. Nat Rev Mol Cell Biol, 2020, 21(9): 542-556.
pmid: 32483315 |
| [64] |
Jones MR, Quinton LJ, Blahna MT, Neilson JR, Fu SN, Ivanov AR, Wolf DA, Mizgerd JP. Zcchc11-dependent uridylation of microRNA directs cytokine expression. Nat Cell Biol, 2009, 11(9): 1157-1163.
pmid: 19701194 |
| [65] |
Yang A, Bofill-De Ros X, Shao TJ, Jiang MJ, Li K, Villanueva P, Dai LS, Gu S. 3′ uridylation confers miRNAs with non-canonical target repertoires. Mol Cell, 2019, 75(3): 511-522.e4.
pmid: 31178353 |
| [66] |
Baumberger N, Baulcombe DC. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA, 2005, 102(33): 11928-11933.
pmid: 16081530 |
| [67] |
Li SB, Liu L, Zhuang XH, Yu Y, Liu XG, Cui X, Ji LJ, Pan ZQ, Cao XF, Mo BX, Zhang FC, Raikhel N, Jiang LW, Chen XM. MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell, 2013, 153(3): 562-574.
pmid: 23622241 |
| [68] |
Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O. Widespread translational inhibition by plant miRNAs and siRNAs. Science, 2008, 320(5880): 1185-1190.
pmid: 18483398 |
| [69] |
Chen XM. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science, 2004, 303(5666): 2022-2025.
pmid: 12893888 |
| [70] |
Sheu-Gruttadauria J, Pawlica P, Klum SM, Wang SN, Yario TA, Schirle Oakdale NT, Steitz JA, MacRae IJ. Structural basis for target-directed microRNA degradation. Mol Cell, 2019, 75(6): 1243-1255.e7.
pmid: 31353209 |
| [71] |
Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell, 2009, 136(2): 215-233.
pmid: 19167326 |
| [72] |
Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng ZP, Zamore PD. Target RNA-directed trimming and tailing of small silencing RNAs. Science, 2010, 328(5985): 1534-1539.
pmid: 20558712 |
| [73] |
Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet, 2007, 39(8): 1033-1037.
pmid: 17643101 |
| [74] |
Todesco M, Rubio-Somoza I, Paz-Ares J, Weigel D. A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet, 2010, 6(7): e1001031.
pmid: 20661442 |
| [75] |
Yan J, Gu YY, Jia XY, Kang WJ, Pan SJ, Tang XQ, Chen XM, Tang GL. Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell, 2012, 24(2): 415-427.
pmid: 22345490 |
| [76] |
Reichel M, Li YJ, Li JY, Millar AA. Inhibiting plant microRNA activity: molecular SPONGEs, target MIMICs and STTMs all display variable efficacies against target microRNAs. Plant Biotechnol J, 2015, 13(7): 915-926.
pmid: 25600074 |
| [77] |
Peng T, Qiao MM, Liu HP, Teotia S, Zhang ZH, Zhao YF, Wang BB, Zhao DJ, Shi LN, Zhang C, Le B, Rogers K, Gunasekara C, Duan HT, Gu YY, Tian L, Nie JF, Qi J, Meng FR, Huang L, Chen QH, Wang ZL, Tang JS, Tang XQ, Lan T, Chen XM, Wei HR, Zhao QZ, Tang GL. A resource for inactivation of microRNAs using short tandem target mimic technology in model and crop plants. Mol Plant, 2018, 11(11): 1400-1417.
pmid: 30243763 |
| [78] |
Li FF, Wang WD, Zhao N, Xiao BG, Cao PJ, Wu XF, Ye CY, Shen EH, Qiu J, Zhu QH, Xie JH, Zhou XP, Fan LJ. Regulation of nicotine biosynthesis by an endogenous target mimicry of microRNA in tobacco. Plant Physiol, 2015, 169(2): 1062-1071.
pmid: 26246450 |
| [79] |
Du QG, Wang K, Zou C, Xu C, Li WX. The PILNCR1- miR399 regulatory module is important for low phosphate tolerance in maize. Plant Physiol, 2018, 177(4): 1743-1753.
pmid: 29967097 |
| [80] |
Wang XB, Yan LX, Li TH, Zhang J, Zhang YJ, Zhang JJ, Lian XD, Zhang HP, Zheng XB, Hou N, Cheng J, Wang W, Zhang LL, Ye X, Li JD, Feng JC, Tan B. The lncRNA1-miR6288b-3p-PpTCP4-PpD2 module regulates peach branch number by affecting brassinosteroid biosynthesis. New Phytol, 2024, 243(3): 1050-1064.
pmid: 38872462 |
| [81] |
Guo AH, Nie HS, Li HJ, Li B, Cheng C, Jiang KY, Zhu SW, Zhao N, Hua JP. The miR3367-lncRNA67-GhCYP724B module regulates male sterility by modulating brassinosteroid biosynthesis and interacting with Aorf27 in Gossypium hirsutum. J Integr Plant Biol, 2025, 67(1): 169-190.
pmid: 39526576 |
| [82] |
Wu HJ, Wang ZM, Wang M, Wang XJ. Widespread long noncoding RNAs as endogenous target mimics for microRNAs in plants. Plant Physiol, 2013, 161(4): 1875-1884.
pmid: 23429259 |
| [83] |
Ivashuta S, Banks IR, Wiggins BE, Zhang YJ, Ziegler TE, Roberts JK, Heck GR. Regulation of gene expression in plants through miRNA inactivation. PLoS One, 2011, 6(6): e21330.
pmid: 21731706 |
| [84] |
Ma X, Liu CY, Gu LF, Mo BX, Cao XF, Chen XM. TarHunter, a tool for predicting conserved microRNA targets and target mimics in plants. Bioinformatics, 2018, 34(9): 1574-1576.
pmid: 29236948 |
| [85] |
Lang PLM, Christie MD, Dogan ES, Schwab R, Hagmann J, van de Weyer AL, Scacchi E, Weigel D. A role for the F-box protein HAWAIIAN SKIRT in plant microRNA function. Plant Physiol, 2018, 176(1): 730-741.
pmid: 29114080 |
| [86] |
Mei J, Jiang N, Ren GD. The F-box protein HAWAIIAN SKIRT is required for mimicry target-induced microRNA degradation in Arabidopsis. J Integr Plant Biol, 2019, 61(11): 1121-1127.
pmid: 30565372 |
| [87] |
Zhang XB, Jayaweera D, Peters JL, Szecsi J, Bendahmane M, Roberts JA, González-Carranza ZH. The Arabidopsis thaliana F-box gene HAWAIIAN SKIRT is a new player in the microRNA pathway. PLoS One, 2017, 12(12): e0189788.
pmid: 29244865 |
| [88] |
Damayanti F, Lombardo F, Masuda JI, Shinozaki Y, Ichino T, Hoshikawa K, Okabe Y, Wang N, Fukuda N, Ariizumi T, Ezura H. Functional disruption of the tomato putative ortholog of HAWAIIAN SKIRT results in facultative parthenocarpy, reduced fertility and leaf morphological defects. Front Plant Sci, 2019, 10: 1234.
pmid: 31681360 |
| [89] |
Borna RS, Murchie EH, Pyke KA, Roberts JA, Gonzalez-Carranza ZH. The rice EP3 and OsFBK1 E3 ligases alter plant architecture and flower development, and affect transcript accumulation of microRNA pathway genes and their targets. Plant Biotechnol J, 2022, 20(2): 297-309.
pmid: 34543503 |
| [90] |
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell, 2011, 146(3): 353-358.
pmid: 21802130 |
| [91] |
de la Mata M, Gaidatzis D, Vitanescu M, Stadler MB, Wentzel C, Scheiffele P, Filipowicz W, Grosshans H. Potent degradation of neuronal miRNAs induced by highly complementary targets. EMBO Rep, 2015, 16(4): 500-511.
pmid: 25724380 |
| [92] |
Pawlica P, Sheu-Gruttadauria J, MacRae IJ, Steitz JA. How complementary targets expose the microRNA 3' end for tailing and trimming during target-directed microRNA degradation. Cold Spring Harb Symp Quant Biol, 2019, 84: 179-183.
pmid: 32019864 |
| [93] |
Han J, LaVigne CA, Jones BT, Zhang H, Gillett F, Mendell JT. A ubiquitin ligase mediates target-directed microRNA decay independently of tailing and trimming. Science, 2020, 370(6523): eabc9546.
pmid: 33184234 |
| [94] |
Shi CY, Kingston ER, Kleaveland B, Lin DH, Stubna MW, Bartel DP. The ZSWIM8 ubiquitin ligase mediates target-directed microRNA degradation. Science, 2020, 370(6523): eabc9359.
pmid: 33184237 |
| [95] |
Liu QK, Wang F, Axtell MJ. Analysis of complementarity requirements for plant microRNA targeting using a Nicotiana benthamiana quantitative transient assay. Plant Cell, 2014, 26(2): 741-753.
pmid: 24510721 |
| [96] |
Park JH, Shin SY, Shin C. Non-canonical targets destabilize microRNAs in human Argonautes. Nucleic Acids Res, 2017, 45(4): 1569-1583.
pmid: 28119422 |
| [97] |
Chatterjee S, Grosshans H. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature, 2009, 461(7263): 546-549.
pmid: 19734881 |
| [98] |
Ramachandran V, Chen XM. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science, 2008, 321(5895): 1490-1492.
pmid: 18787168 |
| [99] |
Wang XY, Wang Y, Dou YC, Chen L, Wang JL, Jiang N, Guo CC, Yao QQ, Wang CZ, Liu L, Yu B, Zheng BL, Chekanova JA, Ma JB, Ren GD. Degradation of unmethylated miRNA/miRNA*s by a DEDDy-type 3′ to 5′ exoribonuclease Atrimmer 2 in Arabidopsis. Proc Natl Acad Sci USA, 2018, 115(28): E6659-E6667.
pmid: 29941559 |
| [100] |
Zhang WP, Murphy C, Sieburth LE. Conserved RNaseII domain protein functions in cytoplasmic mRNA decay and suppresses Arabidopsis decapping mutant phenotypes. Proc Natl Acad Sci USA, 2010, 107(36): 15981-15985.
pmid: 20798041 |
| [101] |
Gy I, Gasciolli V, Lauressergues D, Morel JB, Gombert J, Proux F, Proux C, Vaucheret H, Mallory AC. Arabidopsis FIERY1, XRN2, and XRN3 are endogenous RNA silencing suppressors. Plant Cell, 2007, 19(11): 3451-3461.
pmid: 17993620 |
| [102] |
Gazzani S, Lawrenson T, Woodward C, Headon D, Sablowski R. A link between mRNA turnover and RNA interference in Arabidopsis. Science, 2004, 306(5698): 1046-1048.
pmid: 15528448 |
| [103] |
Souret FF, Kastenmayer JP, Green PJ. AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol Cell, 2004, 15(2): 173-183.
pmid: 15260969 |
| [104] |
Liu Y, Gao WR, Wu SY, Lu L, Chen YQ, Guo JL, Men SZ, Zhang XM. AtXRN4 affects the turnover of chosen miRNA*s in Arabidopsis. Plants (Basel), 2020, 9(3): 362.
pmid: 32182993 |
| [105] |
Elbarbary RA, Miyoshi K, Myers JR, Du PC, Ashton JM, Tian B, Maquat LE. Tudor-SN-mediated endonucleolytic decay of human cell microRNAs promotes G1/S phase transition. Science, 2017, 356(6340): 859-862.
pmid: 28546213 |
| [106] |
Hu P, Zhao HW, Zhu P, Xiao YS, Miao WL, Wang YS, Jin HL. Dual regulation of Arabidopsis AGO2 by arginine methylation. Nat Commun, 2019, 10(1): 844.
pmid: 30783097 |
| [107] |
Barre-Villeneuve C, Laudié M, Carpentier MC, Kuhn L, Lagrange T, Azevedo-Favory J. The unique dual targeting of AGO1 by two types of PRMT enzymes promotes phasiRNA loading in Arabidopsis thaliana. Nucleic Acids Res, 2024, 52(5): 2480-2497.
pmid: 38321923 |
| [108] |
Maji RK, Leisegang MS, Boon RA, Schulz MH. Revealing microRNA regulation in single cells. Trends Genet, 2025, S0168- 9525(24)00317-2.
pmid: 39863489 |
| [109] |
Alberti C, Manzenreither RA, Sowemimo I, Burkard TR, Wang JK, Mahofsky K, Ameres SL, Cochella L. Cell- type specific sequencing of microRNAs from complex animal tissues. Nat Methods, 2018, 15(4): 283-289.
pmid: 29481550 |
| [1] | 杨怀昊, 郑丙莲. 植物小RNA的产生、作用方式、功能及在农业中的应用前景[J]. 遗传, 2025, 47(8): 928-943. |
| [2] | 徐德钰, 周溪, 任玉洁. 基于RNAi的抗病毒免疫[J]. 遗传, 2025, 47(8): 876-884. |
| [3] | 张潇, 于燕, 宁勇, 洪绮雯, 史怀平. MicroRNA促进基因表达研究进展[J]. 遗传, 2025, 47(7): 729-741. |
| [4] | 安赛男, 杨欢淳, 姜姗, 李靖轩, 张根发. 融入生物信息学分析的综合性探究型表观遗传学实验设计与探索[J]. 遗传, 2025, 47(5): 600-608. |
| [5] | 吴岳阳, 周小燕, 吴玉峰, 黄驹. NMD途径功能缺陷对水稻表型及转录组的影响[J]. 遗传, 2024, 46(7): 540-551. |
| [6] | 张译文, 黄琴, 吴艳芸, 孙月, 韦永龙. LIN28A/B在肿瘤发生发展中的作用研究进展[J]. 遗传, 2024, 46(6): 452-465. |
| [7] | 韩伟, 张庆珍, 杨静, 周喆. 适用于降解样本的201个遗传标记检测体系的构建和验证[J]. 遗传, 2024, 46(4): 306-318. |
| [8] | 张傲, 岑山, 李晓宇. N6-腺苷甲基化修饰及其对LINE-1的调控机制[J]. 遗传, 2024, 46(3): 209-218. |
| [9] | 王艳妮, 李佳. 单细胞DNA甲基化测序数据处理流程与分析方法[J]. 遗传, 2024, 46(10): 807-819. |
| [10] | 何山, 赵健, 宋晓峰. N6-甲基腺苷修饰对女性生殖系统功能的影响[J]. 遗传, 2023, 45(6): 472-487. |
| [11] | 宋鹏辉, 马丽娟, 严冬. 外显子拼接复合体塑造m6A表观转录组的形成[J]. 遗传, 2023, 45(6): 464-471. |
| [12] | 汪佳豪, 赵卿尧, 周月玲, 史良玉, 王楚端, 俞英. 基因芯片在畜禽遗传育种中的应用及展望[J]. 遗传, 2023, 45(12): 1114-1127. |
| [13] | 杜文珍, 李元敬, 吴佳玲, 陈思羽, 姜亮, 刘刚, 谢宁. 丝状真菌Podospora anserina AA11家族裂解多糖单加氧酶基因的鉴定和功能研究[J]. 遗传, 2023, 45(12): 1128-1146. |
| [14] | 许梦萱, 周明. 植物RNA聚合酶IV调控DNA甲基化和发育的研究进展[J]. 遗传, 2022, 44(7): 567-580. |
| [15] | 王娟, 杨悦宁, 朴威兰, 金花. 尿苷酸化:一种重要的细胞内RNA监控方式[J]. 遗传, 2022, 44(6): 449-465. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||

www.chinagene.cn
备案号:京ICP备09063187号-4
总访问:,今日访问:,当前在线:
