遗传 ›› 2024, Vol. 46 ›› Issue (4): 319-332.doi: 10.16288/j.yczz.23-312
收稿日期:2023-12-18
修回日期:2024-01-30
出版日期:2024-04-20
发布日期:2024-03-11
通讯作者:
张文清,刘伟
E-mail:hjx_0925@163.com;mczhangwq@scut.edu.cn;liuwei7@scut.edu.cn
作者简介:洪佳馨,本科生,专业方向:生物学。E-mail: hjx_0925@163.com
基金资助:
Jiaxin Hong( ), Song’en Xu, Wenqing Zhang(
), Song’en Xu, Wenqing Zhang( ), Wei Liu(
), Wei Liu( )
)
Received:2023-12-18
Revised:2024-01-30
Published:2024-04-20
Online:2024-03-11
Contact:
Wenqing Zhang, Wei Liu
E-mail:hjx_0925@163.com;mczhangwq@scut.edu.cn;liuwei7@scut.edu.cn
Supported by:摘要:
中性粒细胞发生是一个高度有序且被精密调控的过程,造血相关转录因子在其中起着关键作用。造血相关转录因子通过与其辅因子相互作用或转录因子之间相互作用形成复杂的调控网络,调控网络的异常可导致白血病的发生。目前参与该过程的转录因子有几十种,它们的结构与功能被广泛研究,但对转录因子之间调控关系的研究则相对缺乏。人PU.1和cMYB参与中性粒细胞发生的多个阶段且它们的异常通常与血液疾病相关,但目前对于在体情况下两者之间是否存在调控关系以及如何相互作用尚不明确。本研究利用cMyb过表达(cmybhyper)和Pu.1缺陷(pu.1G242D/G242D)的斑马鱼模型,通过整体原位杂交、qRT-PCR、荧光报告系统以及拯救实验检测中性粒细胞发育过程中Pu.1和cMyb的相互作用关系。结果显示,在pu.1G242D/G242D突变体中,中性粒细胞的cmyb表达量显著升高,而在cmybhyper中,中性粒细胞的pu.1表达量则无明显变化。进一步在 pu.1G242D/G242D 突变体中注射MO (morpholino)来降低cmyb的表达,并使用SB以及BrdU染色检测中性粒细胞的数量和增殖情况,发现cmyb的敲低可以拯救突变体中中性粒细胞异常增生的表型。这些结果表明,在中性粒细胞发育过程中Pu.1可以负调控cMyb的表达量。最后,本研究通过构建多位点突变质粒和荧光报告系统,证实了Pu.1能够直接结合在cmyb启动子+72 bp位点,从而负调控其表达。综上所述,本研究明确了Pu.1可以通过调控cmyb的表达参与中性粒细胞的发育,为理解两者之间调控关系及其在疾病中的作用提供了新的线索。
洪佳馨, 徐颂恩, 张文清, 刘伟. Pu.1和cMyb在斑马鱼中性粒细胞发育中的相互作用[J]. 遗传, 2024, 46(4): 319-332.
Jiaxin Hong, Song’en Xu, Wenqing Zhang, Wei Liu. The interaction of Pu.1 and cMyb in zebrafish neutrophil development[J]. Hereditas(Beijing), 2024, 46(4): 319-332.
表3
cmyb启动子多位点突变质粒构建引物序列"
| 突变位点 | 引物序列(5′→3′) |
|---|---|
| 1 AACTTTCT | F:ATAACGTCTGCTGGTTACTTTTGGTTATTCTGCAATTACG R:CCAAAAGTAACCAGCAGACGTTATCTGTCACAACACACCC |
| 2 AGTTCTT | F:ACAAACTTAAACAAGCTGCTCTACTTTTCCGTCCAGACCTGA R:AAAGTAGAGCAGCTTGTTTAAGTTTGTCTCCGACATGTCCAA |
| 3 CGGAAA | F:TTGACGTCCAGACCTGACCGTGAAATTCAGTAACAAAATGT R:ATTTCACGGTCAGGTCTGGACGTCAAAGTAGAGCAGCTTGT |
| 4 AGGAAC | F:ATGAATTTCACTTTGCGTCGAGTCACAGTGTTACACAATAC R:GTGACTCGACGCAAAGTGAAATTCATAAGCAACCAAGTGGA |
| 5 GCTTTTCCTTTTCCA | F:TGGAAAATGAGGAGCGAAACTGTTGCAACCCAATGAATGC R:GGTTGCAACAGTTTCGCTCCTCATTTTCCAGAAATGTTCC |
| 6 ATTTCCT | F:TTATTAAAATGATTCACAGTCAATTAACGATGACACAGGG R:CCCTGTGTCATCGTTAATTGACTGTGAATCATTTTAATAA |
| 7 CGTTTTCCTTTATGG | F:CCATAAGTGAAGACGTGCATTTAGGAATGGGGAACTTTAA R:CCATTCCTAAATGCACGTCTTCACTTATGGAGAGACAGTT |
| 8 AAGTTCCCCATTCC | F:GGACTGGGACACTTTAAAAACTGCCCCAATCGAAATAATG R:TTGGGGCAGTTTTTAAAGTGTCCCAGTCCTAAATGCACGT |
| 9 ATTTTCT | F:AGAAGCTTCTGCACAGATATTTAGGCGCAACACTGCTGGA R:CGCCTAAATATCTGTGCAGAAGCTTCTTTCTTCTGCGAAG |
| 10 AGGAAGT | F:GAGAGCTTAATATCAGCGCTTCTGCCTCGAAGAGGAGCTG R:CAGCTCCTCTTCGAGGCAGAAGCGCTGATATTAAGCTCTC |
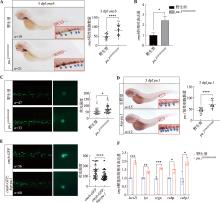
图1
斑马鱼中性粒细胞中Pu.1对cmyb表达的调控作用 A:3 dpf野生型和pu.1G242D/G242D胚胎cmyb mRNA整体原位杂交。红色框显示尾部造血组织,右下角为红色框的局部放大图(200×)。蓝色箭头所示为cmyb阳性信号;B:qRT-PCR检测流式细胞术分选的3 dpf野生型和pu.1G242D/G242D中lyz-dsred阳性中性粒细胞中cmyb的相对表达量;C:在野生型和pu.1G242D/G242D胚胎中分别注射cmyb-GFP质粒后检测3 dpf胚胎中GFP信号强度;D:3 dpf野生型和注射hsp-pu.1质粒的胚胎pu.1 mRNA整体原位杂交。红色框显示尾部造血组织,右下角为红色框的局部放大图(200×)。蓝色箭头所示为pu.1阳性信号;E:在野生型胚胎中单独注射cmyb-GFP质粒和同时注射hsp-pu.1、cmyb-GFP质粒后检测3 dpf胚胎GFP信号强度;F:cmyb靶基因在野生型和pu.1G242D/G242D成鱼肾脏血中的相对表达量。n:样本数;* P<0.05,** P<0.01,*** P<0.001,**** P<0.0001。"

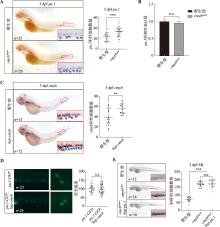
图2
斑马鱼中性粒细胞中cMyb对pu.1表达的调控作用 A:3 dpf野生型和cmybhyper胚胎pu.1 mRNA整体原位杂交。红色框显示尾部造血组织,右下角为红色框的局部放大图(200×)。蓝色箭头所示为pu.1阳性信号;B:qRT-PCR检测流式细胞术分选的3 dpf野生型和cmybhyper 中lyz-dsred阳性细胞中pu.1的相对表达量;C:3 dpf野生型和hsp-cmyb胚胎中cmyb mRNA的整体原位杂交。红色框显示的为尾部造血组织,右下角为红色框的局部放大图(200×),蓝色箭头所示为cmyb阳性信号;D:3 dpf (Tg:pu.1-GFP)与(Tg:pu.1-GFP);hsp-cmyb转基因斑马鱼中GFP信号强度检测;E:3 dpf野生型、cmybhyper和cmybhyper;hsp-pu.1中SB染色,右下角为红色框的局部放大图(200×)。n:样本数;** P<0.01,*** P<0.001,**** P<0.0001,n.s.表示没有统计学差异。"
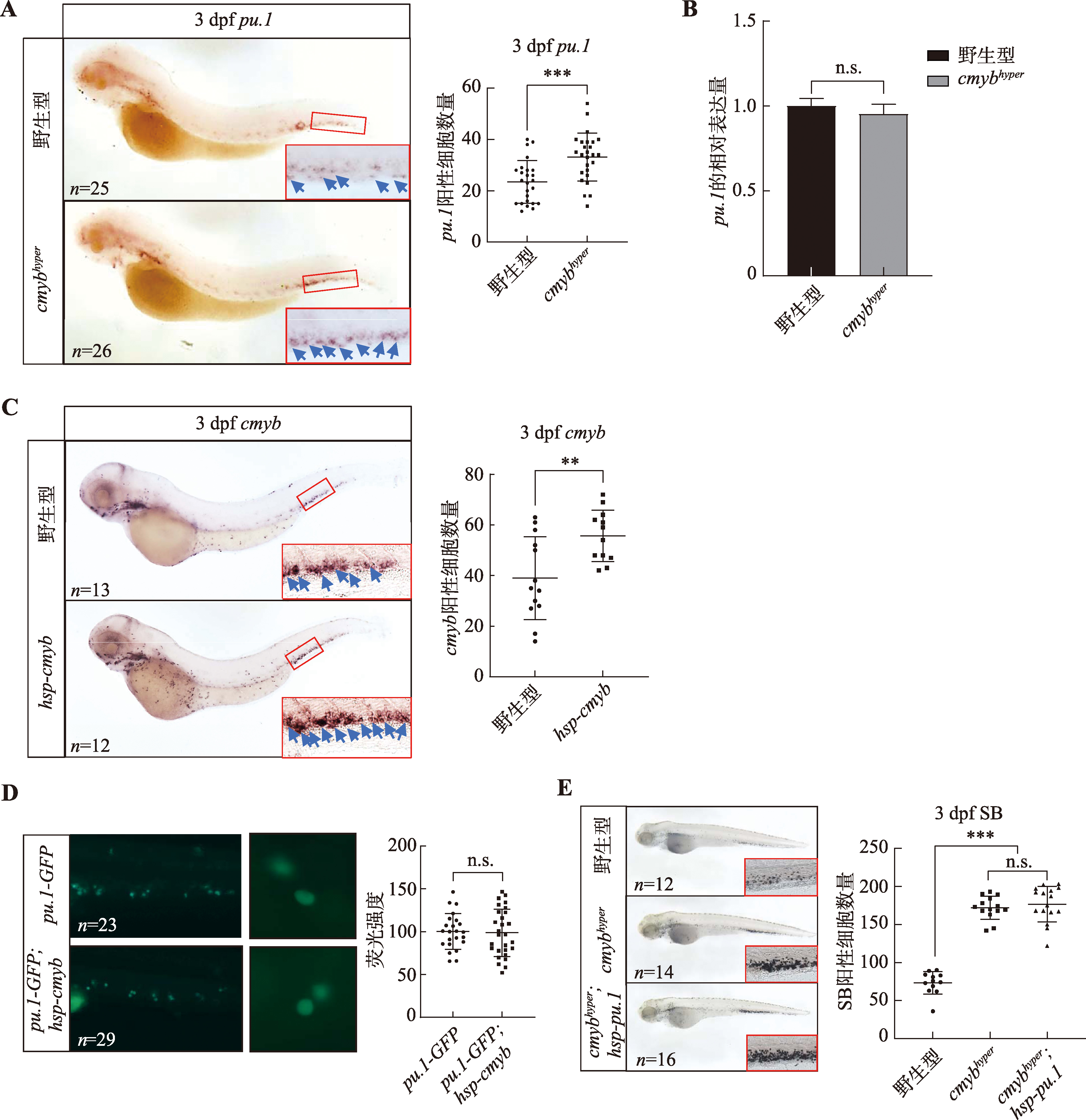
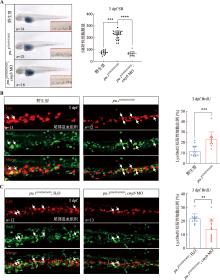
图3
在pu.1G242D/G242D中敲低cmyb对中性粒细胞增殖的影响 A:在1-细胞期的野生型或pu.1G242D/G242D胚胎中分别注射水与cmyb MO,并在3 dpf进行SB染色检测中性粒细胞数量变化;红色框显示的为尾部造血组织,右下角为红色框的局部放大图(200×);B:3 dpf的野生型和pu.1G242D/G242D胚胎尾部造血组织部位Lyz与BrdU的共定位情况,白色箭头所示为共定位的细胞;C:在1-细胞期的pu.1G242D/G242D中分别注射水与cmyb MO,并在3 dpf进行BrdU和Lyz-dsRed免疫荧光染色实验检测尾部造血组织部位增殖的中性粒细胞情况,白色箭头所示为BrdU和Lyz-dsRed双阳性的增殖中性粒细胞。n:样本数;** P<0.01,*** P<0.001,**** P<0.0001。"

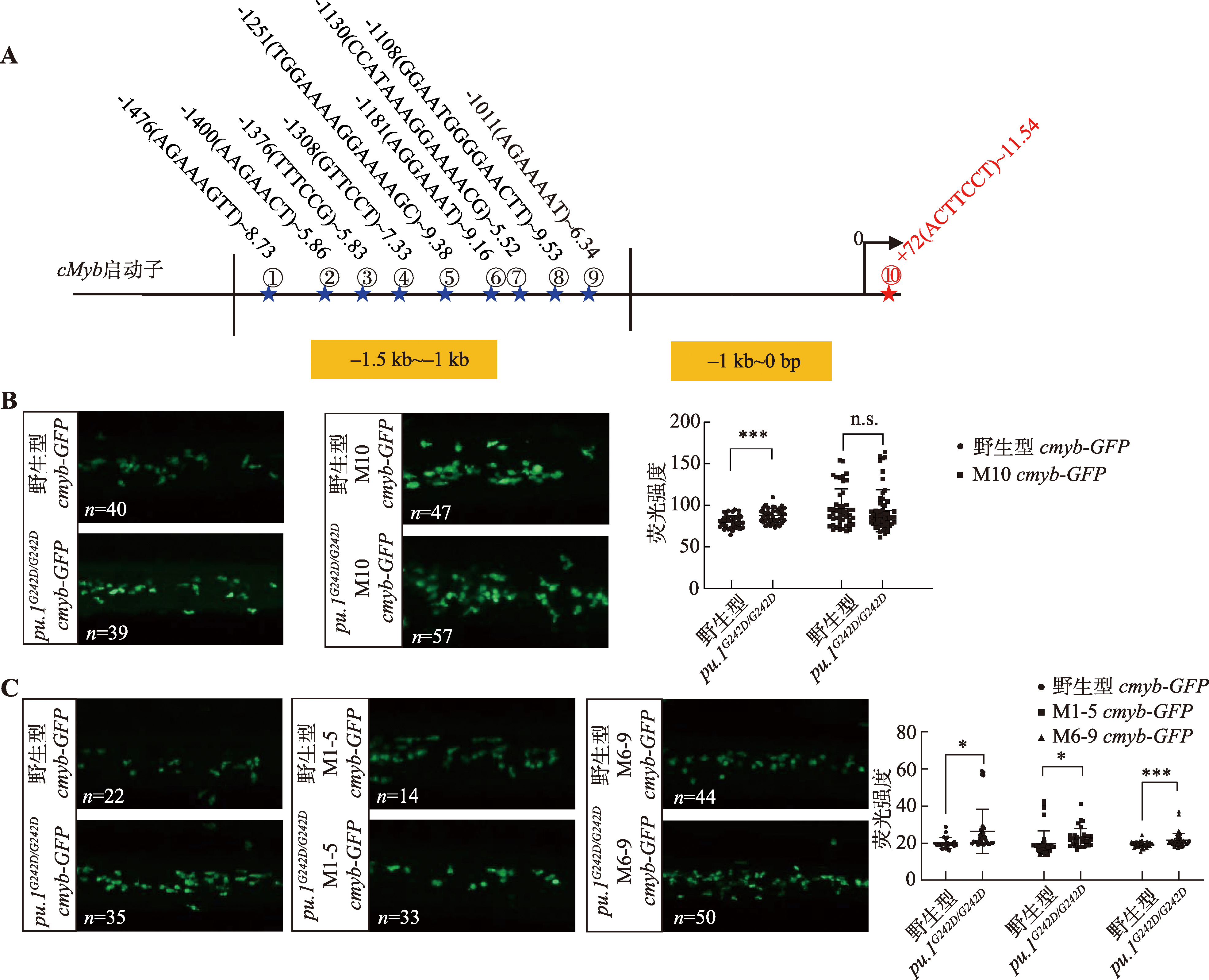
图4
斑马鱼中性粒细胞中cmyb启动子区域Pu.1结合位点的预测与验证 A:JASPAR网站预测cmyb基因启动子上Pu.1的结合位点。蓝色星号指示1~9号位点,红色星号指示10号位点;以+72(ACTTCCT)~11.54为例,+72表示结合位点位于转录起始位点下游72 bp处,(ACTTCCT)表示结合序列,11.54为网站预测给出的评分,评分越高表示结合的可能性越高;B、C:在野生型和pu.1G242D/G242D中分别注射cmyb-GFP、M1-5 cmyb-GFP、M6-9 cmyb-GFP和M10 cmyb-GFP质粒,并检测3 dpf胚胎尾部造血部位GFP信号强度。n:样本数;* P<0.05,*** P<0.001,n.s.表示没有统计学差异。"
| [1] |
Amulic B, Cazalet C, Hayes GL, Metzler KD, Zychlinsky A. Neutrophil function: From mechanisms to disease. Annu Rev Immunol, 2012, 30: 459-489.
doi: 10.1146/annurev-immunol-020711-074942 pmid: 22224774 |
| [2] |
Borregaard N. Neutrophils, from marrow to microbes. Immunity, 2010, 33(5): 657-670.
doi: 10.1016/j.immuni.2010.11.011 pmid: 21094463 |
| [3] |
Galli SJ, Borregaard N, Wynn TA. Phenotypic and functional plasticity of cells of innate immunity: Macrophages, mast cells and neutrophils. Nat Immunol, 2011, 12(11): 1035-1044.
doi: 10.1038/ni.2109 pmid: 22012443 |
| [4] |
Mantovani A, Cassatella MA, Costantini C, Jaillon S. Neutrophils in the activation and regulation of innate and adaptive immunity. Nat Rev Immunol, 2011, 11(8): 519-531.
doi: 10.1038/nri3024 pmid: 21785456 |
| [5] |
Carnevale S, Ceglie ID, Grieco G, Rigatelli A, Bonavita E, Jaillon S. Neutrophil diversity in inflammation and cancer. Front Immunol, 2023, 14: 1180810.
doi: 10.3389/fimmu.2023.1180810 |
| [6] | Lawrence SM, Corriden R, Nizet V. The ontogeny of a neutrophil: Mechanisms of granulopoiesis and homeostasis. Microbiol Mol Biol Rev, 2018, 82(1): e00057-17. |
| [7] |
Antony-Debré I, Paul A, Leite J, Mitchell K, Kim HM, Carvajal LA, Todorova TI, Huang K, Kumar A, Farahat AA, Bartholdy B, Narayanagari SR, Chen JH, Ambesi- Impiombato A, Ferrando AA, Mantzaris I, Gavathiotis E, Verma A, Will B, Boykin DW, Wilson WD, Poon GM, Steidl U. Pharmacological inhibition of the transcription factor pu.1 in leukemia. J Clin Invest, 2017, 127(12): 4297-4313.
doi: 10.1172/JCI92504 pmid: 29083320 |
| [8] |
Anderson KL, Smith KA, Pio F, Torbett BE, Maki RA. Neutrophils deficient in pu.1 do not terminally differentiate or become functionally competent. Blood, 1998, 92(5): 1576-1585.
pmid: 9716585 |
| [9] |
Iwasaki H, Somoza C, Shigematsu H, Duprez EA, Iwasaki-Arai J, Mizuno SI, Arinobu Y, Geary K, Zhang P, Dayaram T, Fenyus ML, Elf S, Chan SS, Kastner P, Huettner CS, Murray R, Tenen DG, Akashi K. Distinctive and indispensable roles of pu.1 in maintenance of hematopoietic stem cells and their differentiation. Blood, 2005, 106(5): 1590-1600.
doi: 10.1182/blood-2005-03-0860 pmid: 15914556 |
| [10] |
Rosenbauer F, Wagner K, Kutok JL, Iwasaki H, Le Beau MM, Okuno Y, Akashi K, Fiering S, Tenen DG. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, pu.1. Nat Genet, 2004, 36(6): 624-630.
pmid: 15146183 |
| [11] |
Sun J, Liu W, Li L, Chen J, Wu M, Zhang Y, Leung AYH, Zhang W, Wen Z, Liao W. Suppression of pu.1 function results in expanded myelopoiesis in zebrafish. Leukemia, 2013, 27(9): 1913-1917.
doi: 10.1038/leu.2013.67 pmid: 23455395 |
| [12] |
Burda P, Curik N, Kokavec J, Basova P, Mikulenkova D, Skoultchi AI, Zavadil J, Stopka T. Pu.1 activation relieves gata-1-mediated repression of cebpa and cbfb during leukemia differentiation. Mol Cancer Res, 2009, 7(10): 1693-1703.
doi: 10.1158/1541-7786.MCR-09-0031 pmid: 19825991 |
| [13] |
Walsh JC, DeKoter RP, Lee HJ, Smith ED, Lancki DW, Gurish MF, Friend DS, Stevens RL, Anastasi J, Singh H. Cooperative and antagonistic interplay between pu.1 and gata-2 in the specification of myeloid cell fates. Immunity, 2002, 17(5): 665-676.
pmid: 12433372 |
| [14] |
Chang HC, Han L, Jabeen R, Carotta S, Nutt SL, Kaplan MH. Pu.1 regulates tcr expression by modulating gata-3 activity. J Immunol, 2009, 183(8): 4887-4894.
doi: 10.4049/jimmunol.0900363 |
| [15] |
Yang Z, Wara-Aswapati N, Chen C, Tsukada J, Auron PE. Nf-il6 (c/ebpβ) vigorously activates il1b gene expression via a spi-1 (pu.1) protein-protein tether. J Biol Chem, 2000, 275(28): 21272-21277.
doi: 10.1074/jbc.M000145200 pmid: 10801783 |
| [16] |
Rekhtman N, Choe KS, Matushansky I, Murray S, Stopka T, Skoultchi AI. Pu.1 and prb interact and cooperate to repress gata-1 and block erythroid differentiation. Mol Cell Biol, 2003, 23(21): 7460-7474.
doi: 10.1128/MCB.23.21.7460-7474.2003 pmid: 14559995 |
| [17] |
Wei F, Zaprazna K, Wang JW, Atchison ML. Pu.1 can recruit bcl6 to DNA to repress gene expression in germinal center b cells. Mol Cell Biol, 2009, 29(17): 4612-4622.
doi: 10.1128/MCB.00234-09 pmid: 19564417 |
| [18] |
Meraro D, Hashmueli S, Koren B, Azriel A, Oumard A, Kirchhoff S, Hauser H, Nagulapalli S, Atchison ML, Levi BZ. Protein-protein and DNA-protein interactions affect the activity of lymphoid-specific ifn regulatory factors. J Immunol, 1999, 163(12): 6468-6478.
pmid: 10586038 |
| [19] |
Zhao XH, Bartholdy B, Yamamoto Y, Evans EK, Alberich- Jordà M, Staber PB, Benoukraf T, Zhang P, Zhang JY, Trinh BQ, Crispino JD, Hoang T, Bassal MA, Tenen DG. Pu.1-c-jun interaction is crucial for pu.1 function in myeloid development. Commun Biol, 2022, 5(1): 961.
doi: 10.1038/s42003-022-03888-7 pmid: 36104445 |
| [20] | Yeamans C, Wang DH, Paz-Priel I, Torbett BE, Tenen DG, Friedman AD. C/ebpα binds and activates the pu.1 distal enhancer to induce monocyte lineage commitment. Blood, 2007, 110(9): 3136-3142. |
| [21] |
Lipsick JS, Wang DM. Transformation by v-myb. Oncogene, 1999, 18(19): 3047-3055.
doi: 10.1038/sj.onc.1202745 pmid: 10378700 |
| [22] |
Clarke ML, Lemma RB, Walton DS, Volpe G, Noyvert B, Gabrielsen OS, Frampton J. Myb insufficiency disrupts proteostasis in hematopoietic stem cells, leading to age-related neoplasia. Blood, 2023, 141(15): 1858-1870.
doi: 10.1182/blood.2022019138 pmid: 36603185 |
| [23] |
Takao S, Forbes L, Uni M, Cheng SY, Pineda JMB, Tarumoto Y, Cifani P, Minuesa G, Chen C, Kharas MG, Bradley RK, Vakoc CR, Koche RP, Kentsis A. Convergent organization of aberrant myb complex controls oncogenic gene expression in acute myeloid leukemia. eLife, 2021, 10: e65905.
doi: 10.7554/eLife.65905 |
| [24] |
Mucenski ML, McLain K, Kier AB, Swerdlow SH, Schreiner CM, Miller TA, Pietryga DW, Scott WJ, Potter SS. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell, 1991, 65(4): 677-689.
doi: 10.1016/0092-8674(91)90099-k pmid: 1709592 |
| [25] |
Sandberg ML, Sutton SE, Pletcher MT, Wiltshire T, Tarantino LM, Hogenesch JB, Cooke MP. C-myb and p 300 regulate hematopoietic stem cell proliferation and differentiation. Dev Cell, 2005, 8(2): 153-166.
doi: 10.1016/j.devcel.2004.12.015 pmid: 15691758 |
| [26] |
Bjerregaard MD, Jurlander J, Klausen P, Borregaard N, Cowland JB. The in vivo profile of transcription factors during neutrophil differentiation in human bone marrow. Blood, 2003, 101(11): 4322-4332.
doi: 10.1182/blood-2002-03-0835 pmid: 12560239 |
| [27] |
Basova P, Pospisil V, Savvulidi F, Burda P, Vargova K, Stanek L, Dluhosova M, Kuzmova E, Jonasova A, Steidl U, Laslo P, Stopka T. Aggressive acute myeloid leukemia in pu.1/p53 double-mutant mice. Oncogene, 2014, 33(39): 4735-4745.
doi: 10.1038/onc.2013.414 pmid: 24121269 |
| [28] |
Liu W, Wu M, Huang Z, Lian J, Chen J, Wang T, Leung AYH, Liao Y, Zhang Z, Liu Q, Yen K, Lin S, Zon LI, Wen Z, Zhang Y, Zhang W. C-myb hyperactivity leads to myeloid and lymphoid malignancies in zebrafish. Leukemia, 2017, 31(1): 222-233.
doi: 10.1038/leu.2016.170 pmid: 27457538 |
| [29] |
Hall C, Flores MV, Storm T, Crosier K, Crosier P. The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. Bmc Dev Biol, 2007, 7: 42.
pmid: 17477879 |
| [30] |
Ward AC, McPhee DO, Condron MM, Varma S, Cody SH, Onnebo SMN, Paw BH, Zon LI, Lieschke GJ. The zebrafish spi1 promoter drives myeloid-specific expression in stable transgenic fish. Blood, 2003, 102(9): 3238-3240.
doi: 10.1182/blood-2003-03-0966 pmid: 12869502 |
| [31] |
Jin H, Li L, Xu J, Zhen FH, Zhu L, Liu PP, Zhang MJ, Zhang WQ, Wen ZL.Runx1 regulates embryonic myeloid fate choice in zebrafish through a negative feedback loop inhibiting pu.1 expression. Blood, 2012, 119(22): 5239-5249.
doi: 10.1182/blood-2011-12-398362 pmid: 22493295 |
| [32] | Westerfield M. The zebrafish book: A guide for the laboratory use of zebrafish (Danio rerio). 4th ed.,University of Oregon Press, Eugene, 1995. |
| [33] | Chitramuthu BP, Bennett HPJ. High resolution whole mount in situ hybridization within zebrafish embryos to study gene expression and function. J Vis Exp, 2013, (80): e50644. |
| [34] |
Lu JA, Huang CY, Lin ZY, Tang Z, Ma N, Huang ZB. The role of the cd99l2 gene on leukocyte interstitial migration in zebrafish. Hereditas(Beijing), 2022, 44(9): 798-809.
doi: 10.16288/j.yczz.22-193 pmid: 36384956 |
| 卢荆澳, 黄春燕, 林芷茵, 唐政, 马宁, 黄志斌. cd99l2基因调控斑马鱼白细胞组织间的迁移机制. 遗传, 2022, 44(9): 798-809. | |
| [35] |
Soza-Ried C, Hess I, Netuschil N, Schorpp M, Boehm T. Essential role of c-myb in definitive hematopoiesis is evolutionarily conserved. Proc Natl Acad Sci USA, 2010, 107(40): 17304-17308.
doi: 10.1073/pnas.1004640107 pmid: 20823231 |
| [36] |
Bellon T, Perrotti D, Calabretta B. Granulocytic differentiation of normal hematopoietic precursor cells induced by transcription factor pu.1 correlates with negative regulation of the c-myb promoter. Blood, 1997, 90(5): 1828-1839.
pmid: 9292515 |
| [37] |
Bies J, Mukhopadhyaya R, Pierce J, Wolff L. Only late, nonmitotic stages of granulocyte differentiation in 32dcl3 cells are blocked by ectopic expression of murine c-myb and its truncated forms. Cell Growth Differ, 1995, 6(1): 59-68.
pmid: 7536440 |
| [38] |
Gotea V, Visel A, Westlund JM, Nobrega MA, Pennacchio LA, Ovcharenko I. Homotypic clusters of transcription factor binding sites are a key component of human promoters and enhancers. Genome Res, 2010, 20(5): 565-577.
doi: 10.1101/gr.104471.109 pmid: 20363979 |
| [39] |
Ezer D, Zabet NR, Adryan B. Homotypic clusters of transcription factor binding sites: a model system for understanding the physical mechanics of gene expression. Comput Struct Biotechnol J, 2014, 10(17): 63-69.
doi: 10.1016/j.csbj.2014.07.005 |
| [40] | Dudek H, Tantravahi RV, Rao VN, Reddy ES, Reddy EP. Myb and ets proteins cooperate in transcriptional activation of the mim-1 promoter. Proc Natl Acad Sci USA, 1992, 89(4): 1291-1295. |
| [41] |
Oelgeschläger M, Nuchprayoon I, Lüscher B, Friedman AD. C/ebp, c-myb, and pu.1 cooperate to regulate the neutrophil elastase promoter. Mol Cell Biol, 1996, 16(9): 4717-4725.
doi: 10.1128/MCB.16.9.4717 pmid: 8756629 |
| [42] |
Ward AC, Loeb DM, Soede-Bobok AA, Touw IP, Friedman AD. Regulation of granulopoiesis by transcription factors and cytokine signals. Leukemia, 2000, 14(6): 973-990.
pmid: 10865962 |
| [1] | 孙飘, 李颖, 刘帆, 王璐. TPI缺乏症斑马鱼模型的构建及分析[J]. 遗传, 2024, 46(3): 232-241. |
| [2] | 李凯伦, 卢荆奥, 陈小辉, 张文清, 刘伟. 尿囊素促进破骨细胞缺陷斑马鱼骨折修复[J]. 遗传, 2023, 45(4): 341-353. |
| [3] | 卢荆澳, 黄春燕, 林芷茵, 唐政, 马宁, 黄志斌. cd99l2基因调控斑马鱼白细胞组织间的迁移机制[J]. 遗传, 2022, 44(9): 798-809. |
| [4] | 郑鹏飞, 谢海波, 朱盼盼, 赵呈天. 斑马鱼神经底板处神经元的分布及特征[J]. 遗传, 2022, 44(6): 510-520. |
| [5] | 张婷婷, 刘峰. 斑马鱼蛋白酪氨酸硫酸化修饰的检测方法研究[J]. 遗传, 2022, 44(2): 178-186. |
| [6] | 贾婷婷, 雷蕾, 吴歆媛, 蔡顺有, 陈艺璇, 薛钰. 二甲双胍对斑马鱼骨骼发育及损伤修复的机制研究[J]. 遗传, 2022, 44(1): 68-79. |
| [7] | 郭佳妮, 刘帆, 王璐. 斑马鱼血液疾病模型及应用[J]. 遗传, 2020, 42(8): 725-738. |
| [8] | 熊凤,谢训卫,潘鲁媛,李阔宇,柳力月,张昀,李玲璐,孙永华. 国家斑马鱼资源中心的资源、技术和服务建设[J]. 遗传, 2018, 40(8): 683-692. |
| [9] | 许璟瑾, 张文娟, 王静怡, 姚丽云, 潘裕添, 欧一新, 薛钰, . 金线莲抑制斑马鱼黑色素形成的活性组分筛选及机理研究[J]. 遗传, 2017, 39(12): 1178-1187. |
| [10] | 刘姗姗, 张翠珍, 彭刚. 饥饿对幼年斑马鱼下丘脑摄食相关性神经肽表达的影响[J]. 遗传, 2016, 38(9): 821-830. |
| [11] | 张峰华,王厚鹏,黄思雨,熊凤,朱作言,孙永华. 两种密码子优化的Cas9编码基因在斑马鱼胚胎中基因敲除效率的比较[J]. 遗传, 2016, 38(2): 144-154. |
| [12] | 顾爱华 严丽锋. 斑马鱼在再生医学研究中的应用及进展[J]. 遗传, 2013, 35(7): 856-866. |
| [13] | 李礼,罗凌飞. 以斑马鱼为模式动物研究器官的发育与再生[J]. 遗传, 2013, 35(4): 421-432. |
| [14] | 徐冉冉 张从伟 曹羽 王强. 缺失mir122抑制斑马鱼肝脏前体细胞向肝细胞分化[J]. 遗传, 2013, 35(4): 488-494. |
| [15] | 沈延 黄鹏 张博. TALEN构建与斑马鱼基因组定点突变的实验方法与流程[J]. 遗传, 2013, 35(4): 533-544. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||

www.chinagene.cn
备案号:京ICP备09063187号-4
总访问:,今日访问:,当前在线:
