遗传 ›› 2025, Vol. 47 ›› Issue (4): 456-475.doi: 10.16288/j.yczz.24-208
何锐( ), 郑秀娟, 王宁宁, 李旭颖, 李明其, 辇士晶, 王克维(
), 郑秀娟, 王宁宁, 李旭颖, 李明其, 辇士晶, 王克维( )
)
收稿日期:2024-09-09
修回日期:2024-11-29
出版日期:2025-04-20
发布日期:2024-12-03
通讯作者:
王克维,教授,博士生导师,研究方向:肝损伤、骨关节炎、细胞凋亡。E-mail: keweiwang0718@163.com作者简介:何锐,硕士研究生,专业方向:流行病与卫生统计学。E-mail: herui55555@163.com
基金资助:
Rui He( ), Xiujuan Zheng, Ningning Wang, Xuying Li, Mingqi Li, Shijing Nian, Kewei Wang(
), Xiujuan Zheng, Ningning Wang, Xuying Li, Mingqi Li, Shijing Nian, Kewei Wang( )
)
Received:2024-09-09
Revised:2024-11-29
Published:2025-04-20
Online:2024-12-03
Supported by:摘要:
PANoptosis是一种新的促炎程序性细胞死亡的方式,参与多种癌症的发生发展过程,但其在肝癌发生发展过程中的机制尚未被阐明。近年来的研究表明,长链非编码RNA (long non-coding RNAs,lncRNAs)在多种癌症的发生发展中起到了关键作用。本研究分别从癌症基因组图谱(The Cancer Genome Atlas,TCGA)数据库和基因表达综合数据库(Gene Expression Omnibus,GEO)数据中检索了肝癌数据集。结合肝癌数据集和前人的研究基础,通过相关性分析得到了PANoptosis相关lncRNAs。一致性聚类分析发现,肝细胞癌患者可以分为两种不同的亚型,即Cluster 1亚型和Cluster 2亚型。与Cluster 2亚型相比,Cluster 1亚型具有更好的预后和更高水平的免疫浸润。然后,对PANoptosis相关lncRNAs进行了Lasso-Cox降维分析,构建了风险评估模型用于预测肝细胞癌患者预后。Kaplan-Meier分析显示,低风险组患者生存率较高,受试者工作特征曲线(receiver operating characteristic curve,ROC)和校准曲线证实了模型具有良好的预测能力。这些发现有助于人们更加深入地理解PANoptosis相关lncRNAs在肝细胞癌发生发展中起到的关键作用,为后续的肝细胞癌治疗提供潜在的生物标志物和治疗靶点。
何锐, 郑秀娟, 王宁宁, 李旭颖, 李明其, 辇士晶, 王克维. 基于生物信息学识别肝细胞癌中的PANoptosis相关lncRNAs并构建预后模型[J]. 遗传, 2025, 47(4): 456-475.
Rui He, Xiujuan Zheng, Ningning Wang, Xuying Li, Mingqi Li, Shijing Nian, Kewei Wang. Identification of PANoptosis-related lncRNAs in hepatocellular carcinoma based on bioinformatics and construction of a prognostic model[J]. Hereditas(Beijing), 2025, 47(4): 456-475.
表1
PANoptosis相关基因集的构建"
| 细胞死亡类型 | 基因 |
|---|---|
| 细胞焦亡 (Pyroptosis) | NLRC4、NLRP1、MEFV、PYCARD、GSDMD、CASP1、CASP5、NLRP3、AIM2、CASP4、CASP12、BAX、IRF1、GSDMB、GSDMC、GSDMA、NLRP6、ELAVL1 |
| 细胞凋亡 (Apoptosis) | CASP10、CASP8、CASP2、APAF1、ZBP1、CASP9、CASP6、CASP3、CASP7、TNFRSF1A、DCN、RNF31、TNFAIP3、MAP3K7、PSTPIP2、ADAR、DIABLO、NLRP9、NAIP、CYLD、STAT1、JAK2、IFNGR1、JAK1、TRADD、TBK1 |
| 坏死性凋亡 (Necroptosis) | RIPK3、RIPK1、MLKL、FADD、FAS、TNF、RBCK1 |
表2
TCGA队列中PANoptosis相关lncRNAs的单因素和多因素Cox回归分析"
| 基因 | 单变量 | 多变量 | ||||
|---|---|---|---|---|---|---|
| HR | 95%CI | P.adj | HR | 95%CI | P.adj | |
| AC091057.1 | 1.18 | 1.06~1.32 | 0.007 | 1.05 | 0.87~1.26 | 0.894 |
| FAM99A | 0.95 | 0.90~0.99 | 0.029 | 0.98 | 0.92~1.05 | 0.894 |
| LINC01703 | 1.18 | 1.06~1.32 | 0.007 | 1.01 | 0.88~1.16 | 0.937 |
| LINC02580 | 0.85 | 0.74~0.96 | 0.014 | 0.93 | 0.79~1.09 | 0.829 |
| AL445524.1 | 1.22 | 1.07~1.39 | 0.007 | 1.13 | 0.95~1.35 | 0.765 |
| LINC01093 | 0.93 | 0.88~0.99 | 0.034 | 0.98 | 0.89~1.07 | 0.894 |
| LINC02499 | 0.91 | 0.85~0.98 | 0.014 | 0.95 | 0.87~1.04 | 0.765 |
| LINC02475 | 1.12 | 1.04~1.20 | 0.007 | 1.08 | 0.99~1.18 | 0.765 |
| AC109322.1 | 1.20 | 1.05~1.38 | 0.012 | 0.97 | 0.80~1.19 | 0.937 |
| AL365361.1 | 0.91 | 0.82~1.00 | 0.049 | 0.94 | 0.82~1.06 | 0.765 |
| AL133338.1 | 1.18 | 1.05~1.33 | 0.012 | 1.11 | 0.96~1.30 | 0.765 |
| FBXL19-AS1 | 1.15 | 1.02~1.3 | 0.029 | 0.99 | 0.84~1.17 | 0.937 |
| AC007773.1 | 1.21 | 1.07~1.36 | 0.007 | 1.03 | 0.85~1.23 | 0.937 |
| AC010864.1 | 1.30 | 1.10~1.55 | 0.007 | 1.13 | 0.90~1.41 | 0.765 |
| AC102953.2 | 1.20 | 1.04~1.38 | 0.014 | 1.00 | 0.84~1.20 | 0.983 |
| AC079174.2 | 1.21 | 1.05~1.40 | 0.012 | 0.93 | 0.75~1.15 | 0.894 |
| AC023593.1 | 1.30 | 1.14~1.49 | <0.001 | 1.13 | 0.92~1.39 | 0.765 |
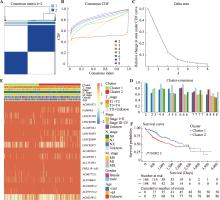
图3
一致性聚类分析基于预后PANoptosis相关lncRNAs将肝癌患者分为两个亚型 A:矩阵热图揭示了一致性分类情况。矩阵的行和列表示的都是样本,当k=2时,其区分度较高。B:一致性累积分布函数(Cumulative Distribution Function,CDF)图展示了k取不同数值时的累积分布函数。C:三角洲面积图(Delta Area Plot)展示了k和k-1相比CDF曲线下面积的相对变化情况。D:聚类共识图展示了不同k值下每个分类的聚类共时值(cluster-consensus value)。该值越高代表稳定性越高。E:分组热图显示了两个亚型患者临床病理特征的关系。F:Kaplan-Meier生存曲线显示两个亚型患者的生存率情况。"
表3
风险分数与临床特征的单因素和多因素COX分析"
| 变量 | 单变量 | 多变量 | ||||
|---|---|---|---|---|---|---|
| HR | 95%CI | P.adj | HR | 95%CI | P.adj | |
| Age | 1.27 | 0.90~1.80 | 0.267 | 1.07 | 0.75~1.54 | 0.757 |
| Gender | 1.23 | 0.86~1.75 | 0.332 | 1.06 | 0.74~1.52 | 0.757 |
| M_stage | 1.27 | 1.06~1.53 | 0.023 | 1.27 | 0.99~1.63 | 0.168 |
| N_stage | 1.23 | 1.03~1.48 | 0.043 | 1.09 | 0.86~1.39 | 0.720 |
| Stage | 1.79 | 1.42~2.27 | <0.001 | 1.16 | 0.79~1.70 | 0.720 |
| T_stage | 2.24 | 1.63~3.08 | <0.001 | 2.11 | 1.33~3.37 | 0.009 |
| Treatment | 1.17 | 0.70~1.97 | 0.589 | 1.49 | 0.90~2.47 | 0.266 |
| Treatment_type | 1.10 | 0.78~1.55 | 0.589 | 1.09 | 0.76~1.55 | 0.757 |
| RiskScore | 3.28 | 2.26~4.75 | <0.001 | 3.65 | 2.49~5.35 | <0.001 |
| [1] |
Vogel A, Meyer T, Sapisochin G, Salem R, Saborowski A. Hepatocellular carcinoma. Lancet, 2022, 400(10360): 1345-1362.
pmid: 36084663 |
| [2] |
Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin, 2022, 72(1): 7-33.
pmid: 35020204 |
| [3] |
Lang H, Sotiropoulos GC, Dömland M, Frühauf NR, Paul A, Hüsing J, Malagó M, Broelsch CE. Liver resection for hepatocellular carcinoma in non-cirrhotic liver without underlying viral hepatitis. Br J Surg, 2004, 92(2): 198-202.
pmid: 15609381 |
| [4] |
Eguchi S, Hara T, Takatsuki M. Liver transplantation for metastatic liver tumors. Hepatol Res, 2017, 47(7): 616-621.
pmid: 28371027 |
| [5] |
Dou JP, Liang P, Yu J. Microwave ablation for liver tumors. Abdom Radiol (NY), 2016, 41(4): 650-658.
pmid: 26880177 |
| [6] |
Bozkurt MF, Salanci BV, Uğur Ö. Intra-arterial radionuclide therapies for liver tumors. Semin Nucl Med, 2016, 46(4): 324-339.
pmid: 27237442 |
| [7] |
Yeung CSY, Chiang CL, Wong NSM, Ha SK, Tsang KS, Ho CHM, Wang B, Lee VWY, Chan MKH, Lee FAS. Palliative liver radiotherapy (RT) for symptomatic hepatocellular carcinoma (HCC). Sci Rep, 2020, 10(1): 1254.
pmid: 31988376 |
| [8] | Han Z, Luo W, Shen J, Xie F, Luo J, Yang X, Pang T, Lv Y, Li Y, Tang X, He J. Non-coding RNAs are involved in tumor cell death and affect tumorigenesis, progression, and treatment: a systematic review. Front Cell Dev Biol, 2024, 12: 1284934. |
| [9] |
Mohammad RM, Muqbil I, Lowe L, Yedjou C, Hsu HY, Lin LT, Siegelin MD, Fimognari C, Kumar NB, Dou QP, Yang HJ, Samadi AK, Russo GL, Spagnuolo C, Ray SK, Chakrabarti M, Morre JD, Coley HM, Honoki K, Fujii H, Georgakilas AG, Amedei A, Niccolai E, Amin A, Ashraf SS, Helferich WG, Yang XJ, Boosani CS, Guha G, Bhakta D, Ciriolo MR, Aquilano K, Chen S, Mohammed SI, Keith WN, Bilsland A, Halicka D, Nowsheen S, Azmi AS. Broad targeting of resistance to apoptosis in cancer. Semin Cancer Biol, 2015, 35(Suppl): S78-S103.
pmid: 25936818 |
| [10] |
Malireddi RKS, Tweedell RE, Kanneganti TD. PANoptosis components, regulation, and implications. Aging (Albany NY), 2020, 12(12): 11163-11164.
pmid: 32575071 |
| [11] |
Jiang MX, Qi L, Li LS, Wu YM, Song DF, Li YJ. Caspase-8: a key protein of cross-talk signal way in "PANoptosis" in cancer. Int J Cancer, 2021, 149(7): 1408-1420.
pmid: 34028029 |
| [12] |
Pandian N, Kanneganti TD. PANoptosis: a unique innate immune inflammatory cell death modality. J Immunol, 2022, 209(9): 1625-1633.
pmid: 36253067 |
| [13] |
Samir P, Malireddi RKS, Kanneganti TD. The PANoptosome: a deadly protein complex driving pyroptosis, apoptosis, and necroptosis (PANoptosis). Front Cell Infect Microbiol, 2020, 10: 238.
pmid: 32582562 |
| [14] |
Ocansey DKW, Qian F, Cai PP, Ocansey S, Amoah S, Qian YC, Mao F. Current evidence and therapeutic implication of PANoptosis in cancer. Theranostics, 2024, 14(2): 640-661.
pmid: 38169587 |
| [15] |
Karki R, Sundaram B, Sharma R, Lee S, Malireddi RKS, Nguyen LN, Christgen S, Zheng M, Wang YQ, Samir P, Neale G, Vogel P, Kanneganti TD. ADAR1 restricts ZBP1-mediated immune response and PANoptosis to promote tumorigenesis. Cell Rep, 2021, 37(3): 109858.
pmid: 34686350 |
| [16] |
Pan HD, Pan JX, Li P, Gao JP. Characterization of PANoptosis patterns predicts survival and immunotherapy response in gastric cancer. Clin Immunol, 2022, 238: 109019.
pmid: 35470064 |
| [17] |
Lin JF, Hu PS, Wang YY, Tan YT, Yu K, Liao K, Wu QN, Li T, Meng Q, Lin JZ, Liu ZX, Pu HY, Ju HQ, Xu RH, Qiu MZ. Phosphorylated NFS1 weakens oxaliplatin-based chemosensitivity of colorectal cancer by preventing PANoptosis. Signal Transduct Target Ther, 2022, 7(1): 54.
pmid: 35221331 |
| [18] |
Wei SY, Chen ZG, Ling XY, Zhang WT, Jiang L. Comprehensive analysis illustrating the role of PANoptosis- related genes in lung cancer based on bioinformatic algorithms and experiments. Front Pharmacol, 2023, 14: 1115221.
pmid: 36874021 |
| [19] |
Zhang JL, Zhao XH, Ma XJ, Yuan ZY, Hu M. KCNQ1OT1 contributes to sorafenib resistance and programmed death‑ligand‑1‑mediated immune escape via sponging miR‑506 in hepatocellular carcinoma cells. Int J Mol Med, 2020, 46(5): 1794-1804.
pmid: 33000204 |
| [20] |
Huarte M. The emerging role of lncRNAs in cancer. Nat Med, 2015, 21(11): 1253-1261.
pmid: 26540387 |
| [21] |
Jarroux J, Morillon A, Pinskaya M. History,discovery, and classification of lncRNAs. Adv Exp Med Biol, 2017, 1008: 1-46.
pmid: 28815535 |
| [22] |
Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet, 2015, 17(1): 47-62.
pmid: 26666209 |
| [23] |
Wang P, Xue YQ, Han YM, Lin L, Wu C, Xu S, Jiang ZP, Xu JF, Liu QY, Cao XT. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science, 2014, 344(6181): 310-313.
pmid: 24744378 |
| [24] |
Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev, 2009, 23(13): 1494-1504.
pmid: 19571179 |
| [25] |
Hao LH, Wu WZ, Xu YK, Chen YF, Meng CZ, Yun JY, Wang XY. lncRNAs-MALAT1: a key participant in the occurrence and development of cancer. Molecules, 2023, 28(5): 2126.
pmid: 36903369 |
| [26] |
Nadhan R, Isidoro C, Song YS, Dhanasekaran DN. lncRNAs and the cancer epigenome: mechanisms and therapeutic potential. Cancer Letters, 2024, 605: 217297.
pmid: 39424260 |
| [27] |
Klec C, Gutschner T, Panzitt K, Pichler M. Involvement of long non-coding RNA HULC (highly up-regulated in liver cancer) in pathogenesis and implications for therapeutic intervention. Expert Opin Ther Targets, 2019, 23(3): 177-186.
pmid: 30678498 |
| [28] |
Li D, Liu XF, Zhou J, Hu J, Zhang DD, Liu J, Qiao YY, Zhan QM. Long noncoding RNA HULC modulates the phosphorylation of YB-1 through serving as a scaffold of extracellular signal-regulated kinase and YB-1 to enhance hepatocarcinogenesis. Hepatology, 2017, 65(5): 1612-1627.
pmid: 28027578 |
| [29] |
Zhao ZB, Gao J, Huang SS. lncRNAs SNHG7 promotes the HCC progression through miR-122-5p/FOXK2 axis. Dig Dis Sci, 2021, 67(3): 925-935.
pmid: 33738672 |
| [30] |
Chen E, Zhou B, Bian SY, Ni WK, Chen Z. The lncRNA B3GALT5-AS1 functions as an HCC suppressor by regulating the miR-934/UFM1 axis. J Oncol, 2021, 2021: 1776432.
pmid: 34721576 |
| [31] | Cheng M, Zhang J, Gao PB, Zhou GQ. Prognostic and predictive value of the hypoxia-associated long non-coding RNA signature in hepatocellular carcinoma. Hereditas(Bejing), 2022, 44(2): 153-173. |
| 程敏, 张静, 曹鹏博, 周钢桥. 缺氧相关长链非编码RNA作为肝癌预后预测标志物的潜在价值. 遗传, 2022, 44(2): 153-173. | |
| [32] |
Deng BH, Wang JY, Yang TY, Deng Z, Yuan JF, Zhang BH, Zhou Z, Chen F, Fang L, Liang CZ, Yan B, Ai YW. TNF and IFNγ-induced cell death requires IRF1 and ELAVL1to promote CASP8 expression. J Cell Biol, 2024, 223(3): e202305026.
pmid: 38319288 |
| [33] |
Wang YQ, Kanneganti TD. From pyroptosis, apoptosis and necroptosis to PANoptosis: a mechanistic compendium of programmed cell death pathways. Comput Struct Biotechnol J, 2021, 19: 4641-4657.
pmid: 34504660 |
| [34] |
Chen XJ, Ye Q, Zhao WX, Chi XQ, Xie CR, Wang XM. RBCK1 promotes hepatocellular carcinoma metastasis and growth by stabilizing RNF31. Cell Death Discov, 2022, 8(1): 334.
pmid: 35869046 |
| [35] |
Wang JK, Huang ZH, Lu HC, Zhang RGY, Feng Q, He AX. A pyroptosis-related gene signature to predict patients’ prognosis and immune landscape in liver hepatocellular carcinoma. Comput Math Methods Med, 2022, 2022: 1258480.
pmid: 35242200 |
| [36] |
Schneider AT, Gautheron J, Feoktistova M, Roderburg C, Loosen SH, Roy S, Benz F, Schemmer P, Büchler MW, Nachbur U, Neumann UP, Tolba R, Luedde M, Zucman-Rossi J, Panayotova-Dimitrova D, Leverkus M, Preisinger C, Tacke F, Trautwein C, Longerich T, Vucur M, Luedde T. RIPK1 suppresses a TRAF2-dependent pathway to liver cancer. Cancer Cell, 2017, 31(1): 94-109.
pmid: 28017612 |
| [37] |
Hellerbrand C, Bumes E, Bataille F, Dietmaier W, Massoumi R, Bosserhoff AK. Reduced expression of CYLD in human colon and hepatocellular carcinomas. Carcinogenesis, 2006, 28(1): 21-27.
pmid: 16774947 |
| [38] |
Li Y, Gan LP, Lu M, Zhang XD, Tong XM, Qi DD, Zhao Y, Ye X. HBx downregulated decorin and decorin-derived peptides inhibit the proliferation and tumorigenicity of hepatocellular carcinoma cells. FASEB J, 2023, 37(4): e22871.
pmid: 36929160 |
| [39] |
Zhang L, Zhang XZ, Zhao W, Xiao XY, Liu SS, Peng QL, Jiang N, Zhou BY. NLRP6-dependent pyroptosis-related lncRNAs predict the prognosis of hepatocellular carcinoma. Front Med (Lausanne), 2022, 9: 760722.
pmid: 35308537 |
| [40] |
Bruix J, Sherman M, American association for the study of liver diseases. Management of hepatocellular carcinoma: an update. Hepatology, 2011, 53(3): 1020-1022.
pmid: 21374666 |
| [41] |
Kuhlmann JB, Blum HE. Locoregional therapy for cholangiocarcinoma. Curr Opin Gastroenterol, 2013, 29(3): 324-328.
pmid: 23337933 |
| [42] |
Banales JM, Marin JJG, Lamarca A, Rodrigues PM, Khan SA, Roberts LR, Cardinale V, Carpino G, Andersen JB, Braconi C, Calvisi DF, Perugorria MJ, Fabris L, Boulter L, Macias RIR, Gaudio E, Alvaro D, Gradilone SA, Strazzabosco M, Marzioni M, Coulouarn C, Fouassier L, Raggi C, Invernizzi P, Mertens JC, Moncsek A, Ilyas SI, Heimbach J, Koerkamp BG, Bruix J, Forner A, Bridgewater J, Valle JW, Gores GJ. Cholangiocarcinoma 2020: the next horizon in mechanisms and management. Nat Rev Gastroenterol Hepatol, 2020, 17(9): 557-588.
pmid: 32606456 |
| [43] |
Carlino MS, Larkin J, Long GV. Immune checkpoint inhibitors in melanoma. Lancet, 2021, 398(10304): 1002-1014.
pmid: 34509219 |
| [44] |
Liu SY, Wu YL. Ongoing clinical trials of PD-1 and PD-L1 inhibitors for lung cancer in China. J Hematol Oncol, 2017, 10(1): 136.
pmid: 28679395 |
| [45] |
Awad MM, Hammerman PS. Durable responses with PD-1 inhibition in lung and kidney cancer and the ongoing search for predictive biomarkers. J Clin Oncol, 2015, 33(18): 1993-1994.
pmid: 25918290 |
| [46] |
Fridman WH, Pagès F, Sautès-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer, 2012, 12(4): 298-306.
pmid: 22419253 |
| [47] |
Pan X, Li CC, Feng JF. The role of lncRNAs in tumor immunotherapy. Cancer Cell Int, 2023, 23(1): 30.
pmid: 36810034 |
| [48] |
Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, Poliakov A, Cao XH, Dhanasekaran SM, Wu YM, Robinson DR, Beer DG, Feng FY, Iyer HK, Chinnaiyan AM. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet, 2015, 47(3): 199-208.
pmid: 25599403 |
| [49] |
Jiang WY, Deng ZL, Dai XZ, Zhao WH. PANoptosis: a new insight into oral infectious diseases. Front Immunol, 2021, 12: 789610.
pmid: 34970269 |
| [50] |
Gao J, Xiong AY, Liu JL, Li XL, Wang JY, Zhang L, Liu Y, Xiong Y, Li GP, He X. PANoptosis: bridging apoptosis, pyroptosis, and necroptosis in cancer progression and treatment. Cancer Gene Ther, 2024, 31(7): 970-983.
pmid: 38553639 |
| [51] |
Malireddi RKS, Karki R, Sundaram B, Kancharana B, Lee S, Samir P, Kanneganti TD. Inflammatory cell death, PANoptosis, mediated by cytokines in diverse cancer lineages inhibits tumor growth. Immunohorizons, 2021, 5(7): 568-580.
pmid: 34290111 |
| [52] | Wan YX, Zhu XY, Zhao Y, Sun N, Jiang TTF, Xu J. Computational dissection of the regulatory mechanisms of aberrant metabolism in remodeling the microenvironment of breast cancer. Hereditas(Beijing), 2024, 46(10): 871-885. |
| 万羽鑫, 朱欣雨, 赵宇, 孙娜, 江天彤妃, 徐娟. 计算解析异常代谢对乳腺癌微环境重塑的调控机制. 遗传, 2024, 46(10): 871-885. | |
| [53] |
Ouyang FZ, Wu RQ, Wei Y, Liu RX, Yang D, Xiao X, Zheng LM, Li B, Lao XM, Kuang DM. Dendritic cell-elicited B-cell activation fosters immune privilege via IL-10 signals in hepatocellular carcinoma. Nat Commun, 2016, 7: 13453.
pmid: 27853178 |
| [54] |
Zhang CY, Liu S, Yang M. Regulatory T cells and their associated factors in hepatocellular carcinoma development and therapy. World J Gastroenterol, 2022, 28(27): 3346-3358.
pmid: 36158267 |
| [55] | Wagner JA, Romee R, Rosario M, Berrien-Elliott MM, Schneider SE, Leong JW, Sullivan RP, Jewell BA, Schappe T, Abdel-Latif S, Ireland AR, Jaishankar D, King JA, Vij R, Fehniger TA. Human CD56bright NK cells acquire potent anti-leukemia functionality following IL-15 priming. Blood, 2016, 128(22): 550. |
| [56] |
Braumüller H, Wieder T, Brenner E, Aßmann S, Hahn M, Alkhaled M, Schilbach K, Essmann F, Kneilling M, Griessinger C, Ranta F, Ullrich S, Mocikat R, Braungart K, Mehra T, Fehrenbacher B, Berdel J, Niessner H, Meier F, Van Den Broek M, Häring HU, Handgretinger R, Quintanilla-Martinez L, Fend F, Pesic M, Bauer J, Zender L, Schaller M, Schulze-Osthoff K, Röcken M. T-helper-1-cell cytokines drive cancer into senescence. Nature, 2013, 494(7437): 361-365.
pmid: 23376950 |
| [57] |
Ohms M, Möller S, Laskay T. An attempt to polarize human neutrophils toward N1 and N2 phenotypes in vitro. Front Immunol, 2020, 11: 532.
pmid: 32411122 |
| [58] |
Mo ML, Ma XY, Luo YH, Tan C, Liu BH, Tang P, Liao Q, Liu S, Yu HP, Huang DP, Zeng XY, Qiu XQ. Liver-specific lncRNA FAM99A may be a tumor suppressor and promising prognostic biomarker in hepatocellular carcinoma. BMC Cancer, 2022, 22(1): 1098.
pmid: 36289466 |
| [59] |
Xu YH, Jia KL, Zhao XH, Li YT, Zhang Z. lncRNAs LINC01703 promotes the proliferation, migration, and invasion of colorectal cancer by activating PI3K/AKT pathway. J Biochem Mol Toxicol, 2023, 38(1): e23594.
pmid: 38050438 |
| [60] |
Wu WJ, Xiao F, Xiong YP, Sun GF, Guo Y, Zhou X, Hu GW, Huang K, Guo H. N6-methyladenosine (m6A)- connected lncRNAs are linked to survival and immune infiltration in glioma patients. Biosci Rep, 2023, 43(5): BSR20222100.
pmid: 37083719 |
| [61] |
Qin YM, Tu X, Huang MF, Ma CF, Huang QQ, Huang QQ, Shu H, Ou C. Novel long noncoding RNAs, LINC01093 and MYLK-AS1, serve as potential diagnostic and prognostic biomarkers or hepatocellular carcinoma. DNA Cell Biol, 2023, 42(8): 488-497.
pmid: 37527208 |
| [62] |
Wang ZX, Xu ZN, Sun HB, Wang Y, Han ZF, Yu Y, Han XL, Yin YY, Xu L. LINC01093 promotes proliferation and invasion of non-small cell lung cancer cells via targeting akt signaling pathway. Eur Rev Med Pharmacol Sci, 2021, 25(7): 2828.
pmid: 33877685 |
| [1] | 古丽米热·阿布都热依木, 陈莹, 唐淑红, 董红, 汪立芹, 吴阳升, 黄俊成, 林嘉鹏. Mfn2减轻内质网应激抑制绵羊卵泡颗粒细胞凋亡的分子机制[J]. 遗传, 2025, 47(3): 342-350. |
| [2] | 杨剑, 石国娟, 彭昂惠, 徐清波, 王睿琪, 薛雷, 喻昕阳, 孙艺昊. Tip60-FOXO调节果蝇JNK信号通路介导的细胞凋亡【已撤稿】[J]. 遗传, 2024, 46(6): 490-501. |
| [3] | 陈昱颖, 张倩, 桂梦会, 冯岚, 曹鹏博, 周钢桥. PTBP1通过调控FGFR2的可变剪接促进肝癌的发展[J]. 遗传, 2024, 46(1): 46-62. |
| [4] | 杨茂艺, 崔潇月, 郑诗棋, 马诗瑶, 郑增长, 邓万燕. 病原体逃逸宿主固有免疫的新策略:抑制细胞焦亡[J]. 遗传, 2023, 45(11): 986-997. |
| [5] | 王翠玲, 刘信燚, 王亚会, 张争, 王治东, 周钢桥. MCM2通过抑制p53信号通路促进胆管癌细胞的增殖、迁移和侵袭[J]. 遗传, 2022, 44(3): 230-244. |
| [6] | 熊婉迪, 徐开宇, 陆林, 李家立. 长链非编码RNA在阿尔茨海默病中的研究进展[J]. 遗传, 2022, 44(3): 189-197. |
| [7] | 程敏, 张静, 曹鹏博, 周钢桥. 缺氧相关长链非编码RNA作为肝癌预后预测标志物的潜在价值[J]. 遗传, 2022, 44(2): 153-167. |
| [8] | 秦中勇, 石晓, 曹平平, 褚鹰, 管蔚, 杨楠, 程禾, 孙玉洁. 细胞凋亡反应中NOXA基因启动子发挥增强子功能调节BCL2基因表达[J]. 遗传, 2020, 42(11): 1110-1121. |
| [9] | 余同露,蔡栋梁,朱根凤,叶晓娟,闵太善,陈红岩,卢大儒,陈浩明. CSN4基因干扰对乳腺癌MDA-MB-231细胞增殖和凋亡的影响[J]. 遗传, 2019, 41(4): 318-326. |
| [10] | 张华伟, 孟星宇, 李连峰, 杨玉莹, 仇华吉. 长链非编码RNA——抗病毒天然免疫应答的新兴调控因子[J]. 遗传, 2018, 40(7): 525-533. |
| [11] | 周瑞,王以鑫,龙科任,蒋岸岸,金龙. LncRNA调控骨骼肌发育的分子机制及其在家养动物中的研究进展[J]. 遗传, 2018, 40(4): 292-304. |
| [12] | 路畅, 黄银花. 动物长链非编码RNA研究进展[J]. 遗传, 2017, 39(11): 1054-1065. |
| [13] | 李静秋, 杨杰, 周平, 乐燕萍, 龚朝辉. 竞争性内源RNA的生物学功能及其调控[J]. 遗传, 2015, 37(8): 756-764. |
| [14] | 赵建元,丁寄葳,米泽云,周金明,魏涛,岑山. HIV-1初始传播病毒Vpr基因遗传变异对诱导G2期阻滞及细胞凋亡的影响[J]. 遗传, 2015, 37(5): 480-486. |
| [15] | 黄小庆,李丹丹,吴娟. 植物长链非编码RNA研究进展[J]. 遗传, 2015, 37(4): 344-359. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||

www.chinagene.cn
备案号:京ICP备09063187号-4
总访问:,今日访问:,当前在线:
