遗传 ›› 2022, Vol. 44 ›› Issue (2): 153-167.doi: 10.16288/j.yczz.21-416
程敏1,3( ), 张静4, 曹鹏博2(
), 张静4, 曹鹏博2( ), 周钢桥1,2,3(
), 周钢桥1,2,3( )
)
收稿日期:2021-12-02
修回日期:2022-01-11
出版日期:2022-02-20
发布日期:2022-01-19
作者简介:程敏,硕士研究生,专业方向:流行病与卫生统计学。E-mail: 基金资助:
Min Cheng1,3( ), Jing Zhang4, Pengbo Cao2(
), Jing Zhang4, Pengbo Cao2( ), Gangqiao Zhou1,2,3(
), Gangqiao Zhou1,2,3( )
)
Received:2021-12-02
Revised:2022-01-11
Published:2022-02-20
Online:2022-01-19
Supported by:摘要:
肝细胞癌(hepatocellular carcinoma, 简称肝癌)是一种常见的恶性肿瘤。缺氧是肝癌等实体肿瘤的一个重要特征,同时也是诱导肿瘤恶性进展的重要因素。然而,肝癌缺氧相关的长链非编码RNA(long non-coding RNA,lncRNA)的鉴定及其在临床生存预后等方面的价值仍未得到系统的研究。本研究旨在通过肝癌转录组的整合分析鉴定肝癌缺氧相关的lncRNA,并评估其在肝癌预后中的价值。基于癌症基因组图谱(The Cancer Genome Atlas, TCGA)计划的肝癌转录组数据的整合分析,初步鉴定到233个缺氧相关的候选lncRNA。进一步筛选具有预后价值的候选者,基于其中12个缺氧相关lncRNA (AC012676.1、PRR7-AS1、AC020915.2、AC008622.2、AC026401.3、MAPKAPK5-AS1、MYG1-AS1、AC015908.3、AC009275.1、MIR210HG、CYTOR和SNHG3)建立了肝癌预后风险模型。Cox比例风险回归分析显示,基于该模型计算的缺氧风险评分作为肝癌患者新的独立预后预测指标,优于传统的临床病理因素。基因集富集分析显示,缺氧风险评分反映了细胞增殖相关通路的活化和脂代谢过程的失活。综上所述,本研究构建了一个基于缺氧相关lncRNA的风险评分模型,可以作为肝癌患者预后预测的候选指标,并初步提示了这些缺氧相关的lncRNA在肝癌防治中的重要作用。
程敏, 张静, 曹鹏博, 周钢桥. 缺氧相关长链非编码RNA作为肝癌预后预测标志物的潜在价值[J]. 遗传, 2022, 44(2): 153-167.
Min Cheng, Jing Zhang, Pengbo Cao, Gangqiao Zhou. Prognostic and predictive value of the hypoxia-associated long non-coding RNA signature in hepatocellular carcinoma[J]. Hereditas(Beijing), 2022, 44(2): 153-167.
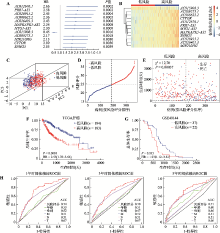
图2
肝癌缺氧相关lncRNA预后模型的构建 A:多变量Cox比例风险回归分析。森林图显示了候选lncRNA的HR(95% CI)和P值。B:热图显示了高风险组患者和低风险组患者中候选lncRNA的表达水平。C:主成分分析显示基于12个候选lncRNA的表达谱可以显著区分低风险组和高风险组肝癌患者。D:缺氧相关lncRNA预后特征的高、低风险肝癌患者风险评分分布。E:肝癌患者的生存时间与基于缺氧相关lncRNA构建的预后特征的风险评分之间的相关性。F:Kaplan-Meier生存曲线显示,TCGA肝癌队列中基于缺氧相关lncRNA构建的预后特征的高风险评分患者的生存时间显著短于低风险评分患者。G:Kaplan-Meier生存曲线显示,GSE40144队列中基于缺氧相关lncRNA构建的预后特征的高风险评分患者的生存时间显著短于低风险评分患者。H:风险模型评分和临床特征的1、3和5年生存预测能力评价。PC1,主成分1(principal component 1);PC2,主成分2(principal component 2);PC3,主成分3(principal component 3);T,肿瘤大小;N,淋巴结转移;M,远端转移。"
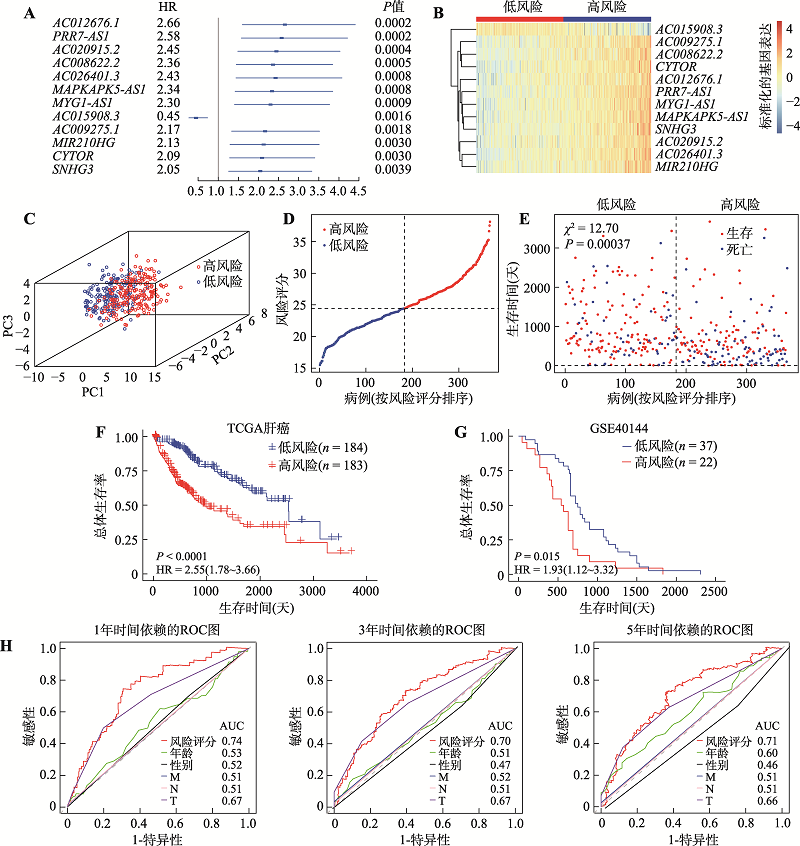

图6
MIR210HG表达水平在肝癌组织中显著上调,且与其基因组拷贝数显著相关 A:MIR210HG在TCGA肝癌癌组织及癌旁组织中的表达水平和MIR210HG高表达组和低表达组的Kaplan-Meier生存曲线。P值通过方差分析进行计算。样本按照MIR210HG的表达中值分为高表达和低表达组。组间差异采用秩和检验。B:MIR210HG在TCGA多种癌症的肿瘤及TCGA和GTEx非肿瘤组织中的表达水平。P值通过方差分析进行计算。*:P<0.01。C:TCGA泛癌队列中基于MIR210HG的表达水平的患者生存分析。HR和P值由Cox比例风险回归分析确定。D:TCGA肝癌组织中MIR210HG基因拷贝数与其表达水平间的相关性。采用Spearman相关分析确定Rho和P值。E:TCGA肝癌组织中MIR210HG表达水平与缺氧评分间的相关性。采用Spearman相关分析确定Rho和P值。F:缺氧条件下MIR210HG和肿瘤缺氧相关基因表达水平显著增加。分别在常氧和氧气浓度2%条件下培养24 h。G:TCGA肝癌组织中MIR210HG表达水平与HIF1A表达水平间的相关性。T,肿瘤样本;N,癌旁对照样本;ACC,肾上腺皮质癌;BLCA,膀胱尿路上皮癌;BRCA,乳腺浸润癌;CESC,宫颈鳞癌和腺癌;CHOL,胆管癌;COAD,结肠癌;DLBC,弥漫性大B细胞淋巴瘤;ESCA,食管癌;GBM,多形成性胶质细胞瘤;HNSC,头颈鳞状细胞癌;KICH,肾嫌色细胞癌;KIRC,肾透明细胞癌;KIRP,肾乳头状细胞癌;LAML,急性髓细胞样白血病;LGG,脑低级别胶质瘤;LIHC,肝细胞肝癌;LUAD,肺腺癌;LUSC,肺鳞癌;MESO,间皮瘤;OV,卵巢浆液性囊腺癌;PAAD,胰腺癌;PCPG,嗜铬细胞瘤和副神经节瘤;PRAD,前列腺癌;READ,直肠腺癌;SARC,肉瘤;SKCM,皮肤黑色素瘤;STAD,胃癌;TGCT,睾丸癌;THCA,甲状腺癌;THYM,胸腺癌;UCEC,子宫内膜癌;UCS,子宫肉瘤;UVM,葡萄膜黑色素瘤。"
附表1
TCGA中肝癌患者的临床信息"
| 特征 | 样本量(n=367) | 比例(%) |
|---|---|---|
| 年龄(岁) | ||
| > 60 | 194 | 52.9 |
| ≤ 60 | 173 | 47.1 |
| 性别 | ||
| 女 | 119 | 32.4 |
| 男 | 248 | 67.6 |
| T分类 | ||
| T1(< 2厘米) | 181 | 49.3 |
| T2(2~5厘米) | 92 | 25.1 |
| T3(≥ 5厘米) | 78 | 21.3 |
| T4(靠近肝脏的血管和/或器官和/或内脏腹膜侵犯) | 13 | 3.5 |
| NA | 3 | 0.8 |
| N分类 | ||
| N0(附近的淋巴结没有癌细胞) | 249 | 67.8 |
| N1(肝脏附近淋巴结的癌细胞) | 4 | 1.1 |
| NA | 114 | 31.1 |
| M分类 | ||
| M0(没有癌细胞已经扩散到肝外的迹象) | 264 | 71.9 |
| M1(癌细胞存在于身体其他器官,如肺部或骨骼) | 3 | 0.8 |
| NA | 100 | 27.3 |
附表2
肿瘤缺氧相关编码基因集"
| MTX1 | ADORA2B | AK3 | ALDOA | ANGPTL4 | C20orf20 | MRPS17 | PGF | PGK1 |
|---|---|---|---|---|---|---|---|---|
| P4HA1 | PFKFB4 | PGAM1 | PVR | SLC16A1 | SLC2A1 | TEAD4 | TPBG | TPI1 |
| GAPD | GMFB | GSS | HES2 | HIG2 | IL8 | KCTD11 | KRT17 | PEDS1 |
| PSMA7 | PSMB7 | PSMD2 | PTGFRN | PYGL | RAN | RNF24 | RNPS1 | RUVBL2 |
| ANLN | VEGF | LOC56901 | S100A3 | B4GALT2 | VEZT | LRP2BP | SIP1 | BCAR1 |
| MGC14560 | SLC6A10 | BMS1L | ANKRD9 | MGC17624 | SLC6A8 | BNIP3 | C14orf156 | MGC2408 |
| HOMER1 | C15orf25 | MIF | SMILE | HSPC163 | CA12 | MRPL14 | SNX24 | IMP-2 |
| NUDT15 | SPTB | KIAA1393 | LDHA | LDLR | MGC2654 | MNAT1 | NDRG1 | NME1 |
| COL4A5 | CORO1C | CTEN | DKFZP564D166 | DPM2 | EIF2S1 | PAWR | PDZK11 | PLAU |
| PPARD | PPP2CZ | PPP4R1 | TFAP2C | TIMM23 | TMEM30B | TPD52L2 | VAPB | XPO5 |
| ANLN | BNC1 | C20orf20 | CA9 | CDKN3 | COL4A6 | DCBLD1 | ENO1 | FAM83B |
| GNAI1 | HIG2 | KCTD11 | KRT17 | LDHA | MPRS17 | P4HA1 | PGAM1 | PGK1 |
| AFARP1 | TUBB2 | LOC149464 | S100A10 | AD-003 | SLCO1B3 | CA9 | CDCA4 | PLEKHG3 |
| ALDOA | FOSL1 | SDC1 | SLC16A1 | SLC2A1 | TPI1 | VEGFA |
附表3
20个在肝癌中具有潜在预后价值的缺氧相关的lncRNAs"
| LncRNA | Rho | HR (95% CI) | P | 类型 |
|---|---|---|---|---|
| AC004816.1 | 0.6042 | 1.83 (1.29~2.60) | 0.00076 | 风险 |
| AC008622.2 | 0.8633 | 2.37 (1.66~3.40) | < 0.0001 | 风险 |
| AC009275.1 | 0.6005 | 1.82 (1.28~2.59) | 0.00083 | 风险 |
| AC012676.1 | 0.7518 | 2.12 (1.49~3.02) | < 0.0001 | 风险 |
| AC015908.3 | -0.7035 | 0.49 (0.35~0.71) | 0.0001 | 保护 |
| AC020915.2 | 0.7523 | 2.12 (1.48~3.04) | < 0.0001 | 风险 |
| AC026401.3 | 0.6128 | 1.85 (1.30~2.63) | 0.00068 | 风险 |
| AC073573.1 | -0.6068 | 0.55 (0.38~0.78) | 0.00093 | 保护 |
| AC114803.1 | 0.9514 | 2.59 (1.81~3.71) | < 0.0001 | 风险 |
| CYTOR | 0.7993 | 2.22 (1.56~3.18) | < 0.0001 | 风险 |
| DANCR | 0.7093 | 2.03 (1.43~2.90) | < 0.0001 | 风险 |
| GIHCG | 0.7331 | 2.08 (1.46~2.97) | < 0.0001 | 风险 |
| MAFG.DT | 0.5451 | 1.72 (1.22~2.44) | 0.0022 | 风险 |
| MAPKAPK5-AS1 | 0.9134 | 2.49 (1.73~2.59) | < 0.0001 | 风险 |
| MIR210HG | 0.7222 | 2.06 (1.44~2.95) | < 0.0001 | 风险 |
| MIR4435.2HG | 0.6980 | 2.01 (1.41~2.86) | 0.00011 | 风险 |
| MYG1-AS1 | 0.5395 | 1.72 (1.21~2.43) | 0.0025 | 风险 |
| PRR7-AS1 | 0.8126 | 2.25 (1.59~3.19) | < 0.0001 | 风险 |
| SNHG3 | 0.6218 | 1.86 (1.31~2.65) | 0.00054 | 风险 |
| TMEM220-AS1 | -0.4673 | 0.63 (0.44~0.89) | 0.0089 | 保护 |
附表4
基于TCGA中肝癌样本风险评分的GSEA结果"
| 基因集 | 基因集大小 | NES | P |
|---|---|---|---|
| GO_RETROGRADE_VESICLE_MEDIATED_TRANSPORT_GOLGI_TO_ER | 77 | 2.34 | 0 |
| GO_NEGATIVE_REGULATION_OF_MITOTIC_CELL_CYCLE | 198 | 1.82 | 0 |
| GO_REGULATION_OF_PROTEASOMAL_UBIQUITIN_DEPENDENT_PROTEIN_CATABOLIC_ PROCESS | 146 | 1.81 | 0 |
| GO_POSITIVE_REGULATION_OF_CELL_CYCLE_PROCESS | 243 | 1.81 | 0 |
| GO_NUCLEAR_CHROMOSOME_SEGREGATION | 218 | 1.81 | 0 |
| GO_TUBULIN_BINDING | 263 | 1.78 | 0 |
| GO_MICROTUBULE_BASED_MOVEMENT | 199 | 1.77 | 0 |
| GO_MICROTUBULE | 397 | 1.77 | 0 |
| GO_SPINDLE_MIDZONE | 27 | 1.81 | 0.0020 |
| GO_REGULATION_OF_CELL_DIVISION | 266 | 1.80 | 0.0021 |
| GO_NEGATIVE_REGULATION_OF_ORGANELLE_ORGANIZATION | 384 | 1.77 | 0.0021 |
| GO_REGULATION_OF_DNA_DEPENDENT_DNA_REPLICATION | 41 | 1.81 | 0.0040 |
| GO_NUCLEAR_UBIQUITIN_LIGASE_COMPLEX | 42 | 1.81 | 0.0041 |
| GO_POSITIVE_REGULATION_OF_G1_S_TRANSITION_OF_MITOTIC_CELL_CYCLE | 24 | 1.78 | 0.0041 |
| GO_CONDENSED_CHROMOSOME | 183 | 1.77 | 0.0043 |
附表4
续"
| 基因集 | 基因集大小 | NES | P |
|---|---|---|---|
| GO_RECOMBINATIONAL_REPAIR | 70 | 1.78 | 0.0063 |
| GO_CONDENSED_CHROMOSOME_CENTROMERIC_REGION | 94 | 1.78 | 0.0064 |
| GO_POSITIVE_REGULATION_OF_MITOTIC_NUCLEAR_DIVISION | 51 | 1.77 | 0.0064 |
| GO_SPINDLE_ASSEMBLY | 68 | 1.81 | 0.0081 |
| GO_HETEROCHROMATIN | 66 | 1.77 | 0.0083 |
| GO_CELL_CYCLE_G2_M_PHASE_TRANSITION | 132 | 1.81 | 0.0084 |
| GO_MITOTIC_CELL_CYCLE_CHECKPOINT | 138 | 1.80 | 0.013 |
| GO_SPINDLE_LOCALIZATION | 38 | 1.78 | 0.014 |
| GO_POSITIVE_REGULATION_OF_CHROMOSOME_SEGREGATION | 25 | 1.77 | 0.014 |
| GO_MEMBRANE_DISASSEMBLY | 46 | 1.79 | 0.021 |
| GO_REGULATION_OF_TELOMERE_MAINTENANCE | 62 | 1.81 | 0.023 |
| GO_CHROMOSOMAL_REGION | 310 | 1.79 | 0.023 |
| GO_SPLICEOSOMAL_COMPLEX | 163 | 1.78 | 0.033 |
| GO_RNA_SPLICING | 335 | 1.79 | 0.038 |
| GO_LIPID_OXIDATION | 68 | -2.15 | 0 |
| GO_MICROBODY | 132 | -2.13 | 0 |
| GO_FATTY_ACID_CATABOLIC_PROCESS | 71 | -2.13 | 0 |
| GO_FATTY_ACID_BETA_OXIDATION | 49 | -2.10 | 0 |
| GO_MICROBODY_PART | 92 | -2.08 | 0 |
| GO_COENZYME_BINDING | 175 | -2.07 | 0 |
| GO_ORGANIC_ACID_CATABOLIC_PROCESS | 202 | -2.03 | 0 |
| GO_FLAVIN_ADENINE_DINUCLEOTIDE_BINDING | 73 | -2.02 | 0 |
| GO_MICROBODY_MEMBRANE | 58 | -2.00 | 0 |
| GO_METHIONINE_METABOLIC_PROCESS | 18 | -1.99 | 0 |
| GO_COFACTOR_BINDING | 258 | -1.97 | 0 |
| GO_MICROBODY_LUMEN | 44 | -1.97 | 0 |
| GO_AMINO_ACID_BETAINE_METABOLIC_PROCESS | 18 | -1.94 | 0 |
| GO_LIPID_HOMEOSTASIS | 107 | -1.89 | 0 |
| GO_BILE_ACID_METABOLIC_PROCESS | 35 | -1.89 | 0 |
| GO_REGULATION_OF_FATTY_ACID_OXIDATION | 27 | -1.84 | 0 |
| GO_BILE_ACID_BIOSYNTHETIC_PROCESS | 20 | -1.82 | 0 |
| GO_CELLULAR_ALDEHYDE_METABOLIC_PROCESS | 83 | -1.82 | 0 |
| GO_ACYLGLYCEROL_HOMEOSTASIS | 29 | -1.82 | 0 |
| GO_MONOOXYGENASE_ACTIVITY | 91 | -1.80 | 0 |
| GO_DRUG_METABOLIC_PROCESS | 39 | -1.78 | 0 |
| GO_POSITIVE_REGULATION_OF_FATTY_ACID_METABOLIC_PROCESS | 33 | -1.78 | 0 |
| GO_NITROGEN_CYCLE_METABOLIC_PROCESS | 15 | -1.75 | 0 |
| GO_STEROID_HYDROXYLASE_ACTIVITY | 31 | -1.70 | 0 |
| GO_IRON_ION_BINDING | 158 | -1.70 | 0 |
| GO_EPOXYGENASE_P450_PATHWAY | 18 | -1.69 | 0 |
附表4
续"
| 基因集 | 基因集大小 | NES | P |
|---|---|---|---|
| GO_OXYGEN_BINDING | 47 | -1.67 | 0 |
| GO_ARACHIDONIC_ACID_MONOOXYGENASE_ACTIVITY | 15 | -1.66 | 0 |
| GO_PEROXISOME_ORGANIZATION | 32 | -2.00 | 0.0020 |
| GO_PROTEIN_DEGLYCOSYLATION | 21 | -1.97 | 0.0020 |
| GO_REGULATION_OF_TRIGLYCERIDE_METABOLIC_PROCESS | 32 | -1.90 | 0.0020 |
| GO_SERINE_FAMILY_AMINO_ACID_METABOLIC_PROCESS | 41 | -1.89 | 0.0020 |
| GO_2_OXOGLUTARATE_METABOLIC_PROCESS | 20 | -1.88 | 0.0020 |
| GO_REGULATION_OF_TRIGLYCERIDE_BIOSYNTHETIC_PROCESS | 17 | -1.88 | 0.0020 |
| GO_CELLULAR_AMINO_ACID_CATABOLIC_PROCESS | 111 | -1.87 | 0.0020 |
| GO_ALPHA_AMINO_ACID_CATABOLIC_PROCESS | 94 | -1.87 | 0.0020 |
| GO_PYRIDOXAL_PHOSPHATE_BINDING | 51 | -1.86 | 0.0020 |
| GO_SULFUR_AMINO_ACID_METABOLIC_PROCESS | 40 | -1.84 | 0.0020 |
| GO_REGULATION_OF_CHOLESTEROL_METABOLIC_PROCESS | 22 | -1.84 | 0.0020 |
| GO_ACYL_COA_DEHYDROGENASE_ACTIVITY | 17 | -1.82 | 0.0020 |
| GO_GLYOXYLATE_METABOLIC_PROCESS | 27 | -1.81 | 0.0020 |
| GO_REGULATION_OF_MITOCHONDRIAL_FISSION | 17 | -1.73 | 0.0020 |
| GO_ENERGY_RESERVE_METABOLIC_PROCESS | 72 | -1.68 | 0.0020 |
| GO_BILE_ACID_TRANSMEMBRANE_TRANSPORTER_ACTIVITY | 15 | -1.66 | 0.0020 |
| GO_SULFUR_AMINO_ACID_BIOSYNTHETIC_PROCESS | 19 | -1.86 | 0.0021 |
| GO_REGULATION_OF_GLUCOSE_METABOLIC_PROCESS | 104 | -1.80 | 0.0021 |
| GO_BLOOD_COAGULATION_INTRINSIC_PATHWAY | 17 | -1.77 | 0.0021 |
| GO_ORGANIC_HYDROXY_COMPOUND_TRANSPORT | 155 | -1.67 | 0.0021 |
| GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_PAIRED_DONORS_WITH_INCORPORATION_OR_REDUCTION_OF_MOLECULAR_OXYGEN_REDUCED_FLAVIN_OR_FLAVOPROTEIN_AS_ONE_DONOR_AND_INCORPORATION_OF_ONE_ATOM_OF_OXYGEN | 26 | -1.65 | 0.0021 |
| GO_FATTY_ACYL_COA_BINDING | 30 | -1.87 | 0.0039 |
| GO_FATTY_ACID_BETA_OXIDATION_USING_ACYL_COA_DEHYDROGENASE | 18 | -1.79 | 0.0040 |
| GO_BILE_ACID_AND_BILE_SALT_TRANSPORT | 31 | -1.76 | 0.0040 |
| GO_GLUCAN_METABOLIC_PROCESS | 58 | -1.75 | 0.0040 |
| GO_REGULATION_OF_LIPID_CATABOLIC_PROCESS | 50 | -1.74 | 0.0040 |
| GO_CELLULAR_LIPID_CATABOLIC_PROCESS | 148 | -1.84 | 0.0041 |
| GO_SMALL_MOLECULE_CATABOLIC_PROCESS | 325 | -1.83 | 0.0041 |
| GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_PAIRED_DONORS_WITH_INCORPORATION_OR_REDUCTION_OF_MOLECULAR_OXYGEN | 149 | -1.75 | 0.0041 |
| GO_RESPONSE_TO_XENOBIOTIC_STIMULUS | 104 | -1.74 | 0.0041 |
| GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_PAIRED_DONORS_WITH_INCORPORATION_OR_REDUCTION_OF_MOLECULAR_OXYGEN_NAD_P_H_AS_ONE_DONOR_AND_INCORPORATION_OF_ONE_ATOM_OF_OXYGEN | 36 | -1.73 | 0.0041 |
| GO_BLOOD_COAGULATION_FIBRIN_CLOT_FORMATION | 24 | -1.73 | 0.0041 |
| GO_DRUG_TRANSMEMBRANE_TRANSPORT | 19 | -1.68 | 0.0041 |
| GO_GLUTAMATE_METABOLIC_PROCESS | 28 | -1.69 | 0.0042 |
| GO_ALPHA_AMINO_ACID_METABOLIC_PROCESS | 226 | -1.80 | 0.0060 |
| GO_PROTEIN_ACTIVATION_CASCADE | 67 | -1.78 | 0.0060 |
附表4
续"
| 基因集 | 基因集大小 | NES | P |
|---|---|---|---|
| GO_ALDEHYDE_DEHYDROGENASE_NAD_ACTIVITY | 19 | -1.72 | 0.0060 |
| GO_ANDROGEN_METABOLIC_PROCESS | 30 | -1.72 | 0.0062 |
| GO_TRANSCRIPTION_FACTOR_ACTIVITY_DIRECT_LIGAND_REGULATED_SEQUENCE_ SPECIFIC_DNA_BINDING | 48 | -1.67 | 0.0062 |
| GO_BRANCHED_CHAIN_AMINO_ACID_METABOLIC_PROCESS | 23 | -1.78 | 0.0079 |
| GO_MANNOSIDASE_ACTIVITY | 15 | -1.76 | 0.0079 |
| GO_REACTIVE_NITROGEN_SPECIES_METABOLIC_PROCESS | 19 | -1.76 | 0.0079 |
| GO_STEROL_TRANSPORT | 50 | -1.69 | 0.0081 |
| GO_GLYCINE_METABOLIC_PROCESS | 17 | -1.68 | 0.0082 |
| GO_MONOCARBOXYLIC_ACID_TRANSMEMBRANE_TRANSPORTER_ACTIVITY | 45 | -1.64 | 0.0083 |
| GO_STEROID_METABOLIC_PROCESS | 232 | -1.78 | 0.0085 |
| GO_S_ADENOSYLMETHIONINE_METABOLIC_PROCESS | 18 | -1.73 | 0.010 |
| GO_CELLULAR_AMINO_ACID_BIOSYNTHETIC_PROCESS | 91 | -1.73 | 0.010 |
| GO_FATTY_ACID_METABOLIC_PROCESS | 287 | -1.70 | 0.010 |
| GO_TRANSMEMBRANE_RECEPTOR_PROTEIN_SERINE_THREONINE_KINASE_ACTIVITY | 17 | -1.68 | 0.010 |
| GO_BENZENE_CONTAINING_COMPOUND_METABOLIC_PROCESS | 24 | -1.68 | 0.010 |
| GO_REGULATION_OF_GLUCONEOGENESIS | 37 | -1.81 | 0.012 |
| GO_ALPHA_AMINO_ACID_BIOSYNTHETIC_PROCESS | 75 | -1.73 | 0.012 |
| GO_MONOCARBOXYLIC_ACID_METABOLIC_PROCESS | 491 | -1.71 | 0.012 |
| GO_THIOESTER_METABOLIC_PROCESS | 83 | -1.70 | 0.012 |
| GO_PROTEIN_LIPID_COMPLEX | 39 | -1.70 | 0.012 |
| GO_PLATELET_DENSE_GRANULE | 20 | -1.71 | 0.013 |
| GO_ARGININE_METABOLIC_PROCESS | 17 | -1.66 | 0.013 |
| GO_CELLULAR_AMINO_ACID_METABOLIC_PROCESS | 328 | -1.75 | 0.014 |
| GO_REGULATION_OF_CELLULAR_KETONE_METABOLIC_PROCESS | 170 | -1.68 | 0.014 |
| GO_TRICARBOXYLIC_ACID_METABOLIC_PROCESS | 37 | -1.82 | 0.016 |
| GO_ASPARTATE_FAMILY_AMINO_ACID_METABOLIC_PROCESS | 55 | -1.80 | 0.016 |
| GO_REGULATION_OF_PROTEIN_ACTIVATION_CASCADE | 34 | -1.76 | 0.016 |
| GO_NEGATIVE_REGULATION_OF_MITOCHONDRION_ORGANIZATION | 39 | -1.71 | 0.016 |
| GO_POSITIVE_REGULATION_OF_TRIGLYCERIDE_METABOLIC_PROCESS | 20 | -1.71 | 0.017 |
| GO_RESPONSE_TO_MERCURY_ION | 15 | -1.69 | 0.017 |
| GO_REGULATION_OF_FATTY_ACID_METABOLIC_PROCESS | 85 | -1.65 | 0.017 |
| GO_STEROL_HOMEOSTASIS | 57 | -1.77 | 0.019 |
| GO_STEROL_METABOLIC_PROCESS | 121 | -1.71 | 0.019 |
| GO_REGULATION_OF_LIPOPROTEIN_LIPASE_ACTIVITY | 15 | -1.64 | 0.019 |
| GO_MITOCHONDRIAL_MATRIX | 406 | -1.84 | 0.020 |
| GO_DICARBOXYLIC_ACID_METABOLIC_PROCESS | 99 | -1.73 | 0.020 |
| GO_PROTEIN_HOMOTETRAMERIZATION | 59 | -1.73 | 0.020 |
| GO_COMPLEMENT_ACTIVATION | 45 | -1.67 | 0.020 |
| GO_POSITIVE_REGULATION_OF_LIPID_CATABOLIC_PROCESS | 25 | -1.69 | 0.021 |
| GO_COFACTOR_METABOLIC_PROCESS | 329 | -1.66 | 0.024 |
附表4
续"
| 基因集 | 基因集大小 | NES | P |
|---|---|---|---|
| GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_THE_CH_CH_GROUP_OF_DONORS | 57 | -1.77 | 0.025 |
| GO_QUATERNARY_AMMONIUM_GROUP_TRANSPORT | 18 | -1.74 | 0.025 |
| GO_SIGNAL_PEPTIDE_PROCESSING | 24 | -1.71 | 0.027 |
| GO_GLUCAN_BIOSYNTHETIC_PROCESS | 25 | -1.68 | 0.027 |
| GO_RETROGRADE_TRANSPORT_VESICLE_RECYCLING_WITHIN_GOLGI | 23 | -1.72 | 0.028 |
| GO_NAD_BINDING | 53 | -1.69 | 0.029 |
| GO_NUCLEOSIDE_BISPHOSPHATE_METABOLIC_PROCESS | 37 | -1.70 | 0.030 |
| GO_ENDOCYTIC_VESICLE_LUMEN | 17 | -1.64 | 0.030 |
| GO_LIGASE_ACTIVITY_FORMING_CARBON_SULFUR_BONDS | 40 | -1.65 | 0.035 |
| GO_TRIGLYCERIDE_RICH_LIPOPROTEIN_PARTICLE | 19 | -1.65 | 0.035 |
| GO_ASPARTATE_FAMILY_AMINO_ACID_BIOSYNTHETIC_PROCESS | 23 | -1.66 | 0.036 |
| GO_COENZYME_A_METABOLIC_PROCESS | 17 | -1.65 | 0.041 |
| GO_2_IRON_2_SULFUR_CLUSTER_BINDING | 21 | -1.65 | 0.044 |
附表5
PCR引物的序列信息"
| 基因 | 正义(5′→3′) | 反义 (5′→3′) |
|---|---|---|
| MIR210HG | TGAGTAGGAACTCTGGGCGA | CCACAATGGGAAGGAGGCAT |
| HIF1A | AGAGGTTGAGGGACGGAGAT | GCACCAAGCAGGTCATAGGT |
| TGFB | GTCTCCCAAGGAAAGGTAGG | CTCTTGAGTCCCTCGCATCC |
| AKT | GCGGCAGGACCGAGC | AGGTCTTGATGTACTCCCCTCG |
| VEGFA | GTCCTGGAGCGTGTACGTTG | CTTCCGGGCTCGGTGATTTA |
| ACTIN | AGAGCCTCGCCTTTGCCGAT | AGAGCCTCGCCTTTGCCGAT |
| [1] |
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin, 2015, 65(2): 87-108.S
doi: 10.3322/caac.21262 |
| [2] |
Yuen VW, Wong CC. Hypoxia-inducible factors and innate immunity in liver cancer. J Clin Invest, 2020, 130(10):5052-5062.
doi: 10.1172/JCI137553 |
| [3] |
Shimamoto K, Tanimoto K, Fukazawa T, Nakamura H, Kanai A, Bono H, Ono H, Eguchi H, Hirohashi N. GLIS1, a novel hypoxia-inducible transcription factor, promotes breast cancer cell motility via activation of WNT5A. Carcinogenesis, 2020, 41(9):1184-1194.
doi: 10.1093/carcin/bgaa010 pmid: 32047936 |
| [4] |
Huang JL, Zheng L, Hu YW, Wang Q. Characteristics of long non-coding RNA and its relation to hepatocellular carcinoma. Carcinogenesis, 2014, 35(3):507-514.
doi: 10.1093/carcin/bgt405 |
| [5] |
Standaert L, Adriaens C, Radaelli E, Van Keymeulen A, Blanpain C, Hirose T, Nakagawa S, Marine JC. The long noncoding RNA Neat1 is required for mammary gland development and lactation. RNA, 2014, 20(12):1844-1849.
doi: 10.1261/rna.047332.114 pmid: 25316907 |
| [6] |
Wang Y, Chen WY, Lian JY, Zhang HB, Yu B, Zhang MJ, Wei FQ, Wu JH, Jiang JX, Jia YS, Mo F, Zhang SR, Liang XD, Mou XZ, Tang JM. The lncRNA PVT1 regulates nasopharyngeal carcinoma cell proliferation via activating the KAT2A acetyltransferase and stabilizing HIF-1α. Cell Death Differ, 2020, 27(2):695-710.
doi: 10.1038/s41418-019-0381-y |
| [7] |
Wang XW, Li L, Zhao KM, Lin QY, Li HY, Xue XT, Ge WJ, He HJ, Liu D, Xie H, Wu Q, Hu Y. A novel LncRNA HITT forms a regulatory loop with HIF-1α to modulate angiogenesis and tumor growth. Cell Death Differ, 2020, 27(4):1431-1446.
doi: 10.1038/s41418-019-0449-8 |
| [8] |
Zeng Z, Xu FY, Zheng H, Cheng P, Chen QY, Ye Z, Zhong JX, Deng SJ, Liu ML, Huang K, Li Q, Li W, Hu YH, Wang F, Wang CY, Zhao G. LncRNA-MTA2TR functions as a promoter in pancreatic cancer via driving deacetylation-dependent accumulation of HIF-1α. Theranostics, 2019, 9(18):5298-5314.
doi: 10.7150/thno.34559 pmid: 31410216 |
| [9] |
Deng SJ, Chen HY, Ye Z, Deng SC, Zhu S, Zeng Z, He C, Liu ML, Huang K, Zhong JX, Xu FY, Li Q, Liu Y, Wang CY, Zhao G. Hypoxia-induced LncRNA-BX111 promotes metastasis and progression of pancreatic cancer through regulating ZEB1 transcription. Oncogene, 2018, 37(44):5811-5828.
doi: 10.1038/s41388-018-0382-1 |
| [10] |
Liang YR, Song XJ, Li YM, Chen B, Zhao WJ, Wang LJ, Zhang HW, Liu Y, Han DW, Zhang N, Ma TT, Wang YJ, Ye FZ, Luo D, Li XY, Yang QF. LncRNA BCRT1 promotes breast cancer progression by targeting miR-1303/PTBP3 axis. Mol Cancer, 2020, 19(1):85.
doi: 10.1186/s12943-020-01206-5 |
| [11] |
Dong L, Cao X, Luo Y, Zhang GQ, Zhang DD. A positive feedback loop of lncRNA DSCR8/miR-98-5p/STAT3/HIF- 1α plays a role in the progression of ovarian cancer. Front Oncol, 2020, 10:1713.
doi: 10.3389/fonc.2020.01713 |
| [12] |
Hua Q, Mi BM, Xu F, Wen J, Zhao L, Liu JJ, Huang G. Hypoxia-induced lncRNA-AC020978 promotes proliferation and glycolytic metabolism of non-small cell lung cancer by regulating PKM2/HIF-1α axis. Theranostics, 2020, 10(11):4762-4778.
doi: 10.7150/thno.43839 |
| [13] |
Winter SC, Buffa FM, Silva P, Miller C, Valentine HR, Turley H, Shah KA, Cox GJ, Corbridge RJ, Homer JJ, Musgrove B, Slevin N, Sloan P, Price P, West CM, Harris AL. Relation of a hypoxia metagene derived from head and neck cancer to prognosis of multiple cancers. Cancer Res, 2007, 67(7):3441-3449.
doi: 10.1158/0008-5472.CAN-06-3322 |
| [14] |
Eustace A, Mani N, Span PN, Irlam JJ, Taylor J, Betts GNJ, Denley H, Miller CJ, Homer JJ, Rojas AM, Hoskin PJ, Buffa FM, Harris AL, Kaanders JHAM, West CML. A 26-gene hypoxia signature predicts benefit from hypoxia- modifying therapy in laryngeal cancer but not bladder cancer. Clin Cancer Res, 2013, 19(17):4879-4888.
doi: 10.1158/1078-0432.CCR-13-0542 pmid: 23820108 |
| [15] |
Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics, 2013, 14:7.
doi: 10.1186/1471-2105-14-7 pmid: 23323831 |
| [16] | Sun ZL, Jing CY, Xiao CT, Li TC. An autophagy-related long non-coding RNA prognostic signature accurately predicts survival outcomes in bladder urothelial carcinoma patients. Aging (Albany NY), 2020, 12(15):15624-15637. |
| [17] |
Steyerberg EW, Vergouwe Y. Towards better clinical prediction models: seven steps for development and an ABCD for validation. Eur Heart J, 2014, 35(29):1925-1931.
doi: 10.1093/eurheartj/ehu207 pmid: 24898551 |
| [18] |
Blanche P, Dartigues JF, Jacqmin-Gadda H. Estimating and comparing time-dependent areas under receiver operating characteristic curves for censored event times with competing risks. Stat Med, 2013, 32(30):5381-5397.
doi: 10.1002/sim.5958 pmid: 24027076 |
| [19] |
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA, 2005, 102(43):15545-15550.
doi: 10.1073/pnas.0506580102 |
| [20] |
Zhang CJ, Zheng YX, Li X, Hu X, Qi F, Luo J. Genome-wide mutation profiling and related risk signature for prognosis of papillary renal cell carcinoma. Ann Transl Med, 2019, 7(18):427.
doi: 10.21037/atm |
| [21] |
Tang FZ, Li CW, Kang BX, Gao G, Li C, Zhang ZM. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res, 2017, 45(W1):W98-W102.
doi: 10.1093/nar/gkx247 |
| [22] |
Kim S, Kang D, Huo ZG, Park Y, Tseng GC. Meta- analytic principal component analysis in integrative omics application. Bioinformatics, 2018, 34(8):1321-1328.
doi: 10.1093/bioinformatics/btx765 |
| [23] | Tang R, Wu JC, Zheng LM, Li ZR, Zhou KL, Zhang ZS, Xu DF, Chen C. Long noncoding RNA RUSC1-AS-N indicates poor prognosis and increases cell viability in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci, 2018, 22(2):388-396. |
| [24] | Torgovnick A, Schumacher B. DNA repair mechanisms in cancer development and therapy. Front Genet, 2015, 6:157. |
| [25] |
Fasanaro P, Greco S, Ivan M, Capogrossi MC, Martelli F. microRNA: emerging therapeutic targets in acute ischemic diseases. Pharmacol Ther, 2010, 125(1):92-104.
doi: 10.1016/j.pharmthera.2009.10.003 |
| [26] |
Feng S, He AB, Wang DL, Kang B. Diagnostic significance of miR-210 as a potential tumor biomarker of human cancer detection: an updated pooled analysis of 30 articles. Onco Targets Ther, 2019, 12:479-493.
doi: 10.2147/OTT |
| [27] |
Kim JH, Park SG, Song SY, Kim JK, Sung JH. Reactive oxygen species-responsive miR-210 regulates proliferation and migration of adipose-derived stem cells via PTPN2. Cell Death Dis, 2013, 4(4):e588.
doi: 10.1038/cddis.2013.117 |
| [28] |
Yang W, Sun T, Cao JP, Liu FJ, Tian Y, Zhu W. Downregulation of miR-210 expression inhibits proliferation, induces apoptosis and enhances radiosensitivity in hypoxic human hepatoma cells in vitro. Exp Cell Res, 2012, 318(8):944-954.
doi: 10.1016/j.yexcr.2012.02.010 pmid: 22387901 |
| [29] |
Ying Q, Liang LH, Guo WJ, Zha RP, Tian Q, Huang SL, Yao J, Ding J, Bao MY, Ge C, Yao M, Li JJ, He XH. Hypoxia-inducible microRNA-210 augments the metastatic potential of tumor cells by targeting vacuole membrane protein 1 in hepatocellular carcinoma. Hepatology, 2011, 54(6):2064-2075.
doi: 10.1002/hep.24614 |
| [30] |
Wu H, Wang T, Liu YQ, Li X, Xu SL, Wu CT, Zou HB, Cao MF, Jin GX, Lang JY, Wang B, Liu BH, Luo XL, Xu C. Mitophagy promotes sorafenib resistance through hypoxia-inducible ATAD3A dependent axis. J Exp Clin Cancer Res, 2020, 39(1):274.
doi: 10.1186/s13046-020-01768-8 |
| [31] |
Ebright RY, Zachariah MA, Micalizzi DS, Wittner BS, Niederhoffer KL, Nieman LT, Chirn B, Wiley DF, Wesley B, Shaw B, Nieblas-Bedolla E, Atlas L, Szabolcs A, Iafrate AJ, Toner M, Ting DT, Brastianos PK, Haber DA, Maheswaran S. HIF1A signaling selectively supports proliferation of breast cancer in the brain. Nat Commun, 2020, 11(1):6311.
doi: 10.1038/s41467-020-20144-w |
| [32] |
Lin YT, Wu KJ. Epigenetic regulation of epithelial- mesenchymal transition: focusing on hypoxia and TGF-β signaling. J Biomed Sci, 2020, 27(1):39.
doi: 10.1186/s12929-020-00632-3 |
| [33] |
Wang XX, Che XF, Yu Y, Cheng Y, Bai M, Yang ZC, Guo QQ, Xie XC, Li DN, Guo M, Hou KZ, Guo WD, Qu XJ, Cao L. Hypoxia-autophagy axis induces VEGFA by peritoneal mesothelial cells to promote gastric cancer peritoneal metastasis through an integrin α5-fibronectin pathway. J Exp Clin Cancer Res, 2020, 39(1):221.
doi: 10.1186/s13046-020-01703-x |
| [34] |
Ramapriyan R, Caetano MS, Barsoumian HB, Mafra ACP, Zambalde EP, Menon H, Tsouko E, Welsh JW, Cortez MA. Altered cancer metabolism in mechanisms of immunotherapy resistance. Pharmacol Ther, 2019, 195:162-171.
doi: 10.1016/j.pharmthera.2018.11.004 |
| [35] |
Chen BW, Zhou Y, Wei T, Wen L, Zhang YB, Shen SC, Zhang J, Ma T, Chen W, Ni L, Wang Y, Bai XL, Liang TB. lncRNA-POIR promotes epithelial-mesenchymal transition and suppresses sorafenib sensitivity simultaneously in hepatocellular carcinoma by sponging miR-182-5p. J Cell Biochem, 2021, 122(1):130-142.
doi: 10.1002/jcb.v122.1 |
| [36] |
Sun Z, Xue SL, Zhang MY, Xu H, Hu XM, Chen SH, Liu YY, Guo MZ, Cui HM. Aberrant NSUN2-mediated m(5)C modification of H19 lncRNA is associated with poor differentiation of hepatocellular carcinoma. Oncogene, 2020, 39(45):6906-6919.
doi: 10.1038/s41388-020-01475-w |
| [37] |
Cai HY, Zhang Y, Zhang HY, Cui C, Li CH, Lu SC. Prognostic role of tumor mutation burden in hepatocellular carcinoma after radical hepatectomy. J Surg Oncol, 2020, 121(6):1007-1014.
doi: 10.1002/jso.v121.6 |
| [38] |
Maleki Vareki S. High and low mutational burden tumors versus immunologically hot and cold tumors and response to immune checkpoint inhibitors. J Immunother Cancer, 2018, 6(1):157.
doi: 10.1186/s40425-018-0479-7 pmid: 30587233 |
| [39] |
Liu GM, Zeng HD, Zhang CY, Xu JW. Identification of a six-gene signature predicting overall survival for hepatocellular carcinoma. Cancer Cell Int, 2019, 19:138.
doi: 10.1186/s12935-019-0858-2 |
| [40] |
Li XY, Jin F, Li Y. A novel autophagy-related lncRNA prognostic risk model for breast cancer. J Cell Mol Med, 2021, 25(1):4-14.
doi: 10.1111/jcmm.v25.1 |
| [41] |
Li XY, Li Y, Yu XM, Jin F. Identification and validation of stemness-related lncRNA prognostic signature for breast cancer. J Transl Med, 2020, 18(1):331.
doi: 10.1186/s12967-020-02497-4 |
| [42] |
Jin LP, Li CY, Liu T, Wang L. A potential prognostic prediction model of colon adenocarcinoma with recurrence based on prognostic lncRNA signatures. Hum Genomics, 2020, 14(1):24.
doi: 10.1186/s40246-020-00270-8 |
| [43] |
Niklasson CU, Fredlund E, Monni E, Lindvall JM, Kokaia Z, Hammarlund EU, Bronner ME, Mohlin S. Hypoxia inducible factor-2α importance for migration, proliferation, and self-renewal of trunk neural crest cells. Dev Dyn, 2021, 250(2):191-236.
doi: 10.1002/dvdy.v250.2 |
| [44] |
Liu OH, Kiema M, Beter M, Ylä-Herttuala S, Laakkonen JP, Kaikkonen MU. Hypoxia-mediated regulation of histone demethylases affects angiogenesis-associated functions in endothelial cells. Arterioscler Thromb Vasc Biol, 2020, 40(11):2665-2677.
doi: 10.1161/ATVBAHA.120.315214 |
| [45] |
Renfrow JJ, Soike MH, West JL, Ramkissoon SH, Metheny-Barlow L, Mott RT, Kittel CA, D'agostino Jr RB, Tatter SB, Laxton AW, Frenkel MB, Hawkins GA, Herpai D, Sanders S, Sarkaria JN, Lesser GJ, Debinski W, Strowd RE. Attenuating hypoxia driven malignant behavior in glioblastoma with a novel hypoxia-inducible factor 2 alpha inhibitor. Sci Rep, 2020, 10(1):15195.
doi: 10.1038/s41598-020-72290-2 pmid: 32938997 |
| [46] |
Yu AJ, Zhao LW, Kang QM, Li J, Chen K, Fu H. Transcription factor HIF1α promotes proliferation, migration, and invasion of cholangiocarcinoma via long noncoding RNA H19/microRNA-612/Bcl-2 axis. Transl Res, 2020, 224:26-39.
doi: 10.1016/j.trsl.2020.05.010 |
| [47] |
Wang L, Sun LK, Liu RY, Mo HY, Niu YS, Chen TX, Wang YF, Han SS, Tu KS, Liu QG. Long non-coding RNA MAPKAPK5-AS1/PLAGL2/HIF-1α signaling loop promotes hepatocellular carcinoma progression. J Exp Clin Cancer Res, 2021, 40(1):72.
doi: 10.1186/s13046-021-01868-z |
| [48] | Hu B, Yang XB, Yang X, Sang XT. LncRNA CYTOR affects the proliferation, cell cycle and apoptosis of hepatocellular carcinoma cells by regulating the miR-125b-5p/KIAA1522 axis. Aging (Albany NY), 2020, 13(2):2626-2639. |
| [49] |
Wei F, Wang Y, Zhou Y, Li Y. Long noncoding RNA CYTOR triggers gastric cancer progression by targeting miR-103/RAB10. Acta Biochim Biophys Sin (Shanghai), 2021, 53(8):1044-1054.
doi: 10.1093/abbs/gmab071 |
| [50] |
Yang JL, Ma Q, Zhang MM, Zhang WF. LncRNA CYTOR drives L-OHP resistance and facilitates the epithelial- mesenchymal transition of colon carcinoma cells via modulating miR-378a-5p/SERPINE1. Cell Cycle, 2021, 20(14):1415-1430.
doi: 10.1080/15384101.2021.1934626 |
| [51] |
Wang Y, Li WJ, Chen XY, Li Y, Wen PH, Xu F. MIR210HG predicts poor prognosis and functions as an oncogenic lncRNA in hepatocellular carcinoma. Biomed Pharmacother, 2019, 111:1297-1301.
doi: S0753-3322(18)37945-9 pmid: 30841443 |
| [52] |
He ZY, Dang J, Song AL, Cui X, Ma ZJ, Zhang ZT. Identification of LINC01234 and MIR210HG as novel prognostic signature for colorectal adenocarcinoma. J Cell Physiol, 2019, 234(5):6769-6777.
doi: 10.1002/jcp.v234.5 |
| [53] |
Ma J, Kong FF, Yang D, Yang H, Wang CC, Cong R, Ma XX. LncRNA MIR210HG promotes the progression of endometrial cancer by sponging miR-337-3p/137 via the HMGA2-TGF-β/Wnt pathway. Mol Ther Nucleic Acids, 2021, 24:905-922.
doi: 10.1016/j.omtn.2021.04.011 |
| [54] | Bu L, Zhang LB, Tian M, Zheng ZB, Tang HJ, Yang QJ. LncRNA MIR210HG facilitates non-small cell lung cancer progression through directly regulation of miR-874/STAT3 axis. Dose Response, 2020, 18(3):1559325820918052. |
| [55] | Wang AH, Jin CH, Cui GY, Li HY, Wang Y, Yu JJ, Wang RF, Tian XY. MIR210HG promotes cell proliferation and invasion by regulating miR-503-5p/TRAF4 axis in cervical cancer. Aging (Albany NY), 2020, 12(4):3205-3217. |
| [56] |
Yu TZ, Li GP, Wang CG, Gong GQ, Wang LW, Li CY, Chen Y, Wang XL. MIR210HG regulates glycolysis, cell proliferation, and metastasis of pancreatic cancer cells through miR-125b-5p/HK2/PKM2 axis. RNA Biol, 2021, 18(12):2513-2530.
doi: 10.1080/15476286.2021.1930755 |
| [57] |
Du Y, Wei N, Ma RL, Jiang SH, Song D. Long noncoding RNA MIR210HG promotes the warburg effect and tumor growth by enhancing HIF-1α translation in triple-negative breast cancer. Front Oncol, 2020, 10:580176.
doi: 10.3389/fonc.2020.580176 |
| [1] | 程芙蓉, 宋文煜, 曹鹏博, 周钢桥. 失巢凋亡相关转录特征预测肝癌预后和免疫微环境的潜在价值[J]. 遗传, 2025, 47(7): 768-785. |
| [2] | 何锐, 郑秀娟, 王宁宁, 李旭颖, 李明其, 辇士晶, 王克维. 基于生物信息学识别肝细胞癌中的PANoptosis相关lncRNAs并构建预后模型[J]. 遗传, 2025, 47(4): 456-475. |
| [3] | 陈昱颖, 张倩, 桂梦会, 冯岚, 曹鹏博, 周钢桥. PTBP1通过调控FGFR2的可变剪接促进肝癌的发展[J]. 遗传, 2024, 46(1): 46-62. |
| [4] | 熊婉迪, 徐开宇, 陆林, 李家立. 长链非编码RNA在阿尔茨海默病中的研究进展[J]. 遗传, 2022, 44(3): 189-197. |
| [5] | 雷常贵, 贾学渊, 孙文靖. 基于癌症基因组图谱计划多组学数据构建胶质母细胞瘤六基因预后模型[J]. 遗传, 2021, 43(7): 665-679. |
| [6] | 骆红波, 曹鹏博, 周钢桥. DNA甲基化驱动的转录表达特征作为肝癌预后预测标志物的价值[J]. 遗传, 2020, 42(8): 775-787. |
| [7] | 张华伟, 孟星宇, 李连峰, 杨玉莹, 仇华吉. 长链非编码RNA——抗病毒天然免疫应答的新兴调控因子[J]. 遗传, 2018, 40(7): 525-533. |
| [8] | 邹俊遐, 陈科. 缺氧诱导因子(HIFs)在肾癌发生中的作用及其分子机制[J]. 遗传, 2018, 40(5): 341-356. |
| [9] | 周瑞,王以鑫,龙科任,蒋岸岸,金龙. LncRNA调控骨骼肌发育的分子机制及其在家养动物中的研究进展[J]. 遗传, 2018, 40(4): 292-304. |
| [10] | 路畅, 黄银花. 动物长链非编码RNA研究进展[J]. 遗传, 2017, 39(11): 1054-1065. |
| [11] | 李静秋, 杨杰, 周平, 乐燕萍, 龚朝辉. 竞争性内源RNA的生物学功能及其调控[J]. 遗传, 2015, 37(8): 756-764. |
| [12] | 黄小庆,李丹丹,吴娟. 植物长链非编码RNA研究进展[J]. 遗传, 2015, 37(4): 344-359. |
| [13] | 杨峰, 易凡, 曹慧青, 梁子才, 杜权. 长链非编码RNA研究进展[J]. 遗传, 2014, 36(5): 456-468. |
| [14] | 李灵, 宋旭. 长链非编码RNA在生物体中的调控作用[J]. 遗传, 2014, 36(3): 228-236. |
| [15] | 樊春燕, 魏强, 郝志强, 李广林. miRNAs调控lincRNAs的生物信息学预测与功能分析[J]. 遗传, 2014, 36(12): 1226-1234. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||

www.chinagene.cn
备案号:京ICP备09063187号-4
总访问:,今日访问:,当前在线:
