遗传 ›› 2022, Vol. 44 ›› Issue (3): 216-229.doi: 10.16288/j.yczz.21-260
收稿日期:2021-07-19
修回日期:2021-11-11
出版日期:2022-03-20
发布日期:2022-01-27
作者简介:付孟,在读硕士研究生,专业方向:动物学。E-mail: 2461849250@qq.com
基金资助:Received:2021-07-19
Revised:2021-11-11
Published:2022-03-20
Online:2022-01-27
Supported by:摘要:
在家犬(Canis lupus familiaris)、牛(Bos taurus)、猪(Sus scrofa domesticus)、绵羊(Ovis aries)、山羊(Capra hircus)等家养动物之后,家马(Equus caballus)才被人类成功驯化。虽然驯化历史很短,但其对人类社会文明发展变革的影响却最大。家马出色的负重移动能力使人类社会由固定的农耕模式向移动分享模式过渡,使历史发展进入了快车道,因此其起源驯化历史一直备受关注。然而由于家马直系同源野生种早已灭绝,加之现代品种化培育引起遗传多样性骤减,使得相关研究长期争议不断。随着测序技术的不断发展和古代样品的逐步丰富,目前对家马起源驯化过程、群体遗传结构等方面的研究越来越深入。本文从核基因、mtDNA、Y染色体、古DNA等不同层面综述了家马起源与驯化历史方面的研究进展,从品种分化状况、群体演化特征等方面讨论了现代家马品种的群体遗传结构,最后总结了马匹毛色、速度、体型等重要表型性状的遗传基础,以期为今后家马的起源驯化研究、种质资源保护与开发、品种优化方向、现代马业发展等方面提供参考。
付孟, 李艳. 家马的起源历史与品种驯化特征[J]. 遗传, 2022, 44(3): 216-229.
Meng Fu, Yan Li. The origin and domestication history of domestic horses and the domestication characteristics of breeds[J]. Hereditas(Beijing), 2022, 44(3): 216-229.
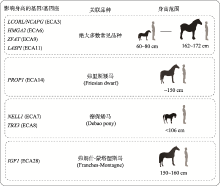
图1
影响家马体高的相关候选基因 体高指由肩隆最高点到地面的距离。体型差异与LASPI、LCORL/NCAPG、HMGA2和ZFAT等4个基因座相关的品种有:阿克哈-塔克马(Akhal-teke horse)、美国迷你马(America Miniature horse)、安达卢西亚马(Andalusian horse)、阿拉伯马(Arabian horse)、阿登纳斯马(Ardennais horse)、比利时马(Belgian horse)、布拉班特马(Braban horse)、里海马(Caspian horse)、克莱兹代尔马(Clydesdale horse)、埃克斯穆尔马(Exmoor horse)、法拉贝拉马(Falabella horse)、费尔矮马(Fell pony)、芬兰马(Finnish horse)、弗朗什-蒙塔涅斯马(Franches-Montagne horse)、法国快步马(French Trotter horse)、荷兰马(Friesian horse)、汉诺威马(Hanoverian horse)、冰岛马(Icelandic horse)、巴西马(Mangalarga Paulista horse)、新福里斯特小型马(New Forest pony)、北方瑞典马(North Swedish horse)、挪威峡湾马(Norwegian Fjord horse)、花马(Paint horse)、佩尔什马(Percheron horse)、秘鲁马(Peruvian Paso horse)、波多黎各小马(Puerto Rican Paso pony horse)、夸特马(Quarter horse)、(Saddlebred horse)、设德兰矮马(Shetland pony)、希尔马(Shire horse)、标准马(Standardbred horse)、萨福克矮马(Suffolk punck horse)、瑞士温血马(Swiss warmblood horse)、田纳西走马(Tennessee walking horse)、纯血马(Thoroughbred horse)、图瓦马(Tuva horse)、威尔士山地小型马(Welsh mountain pony horse)、威尔小马(Welsh pony horse)等。根据参考文献[30, 31, 58~60]绘制。"

表1
马的品种分类方法"
| | 生物学分类 | 畜牧学分类 | 冷热血统分类 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 草原种 | 沙漠种 | 山地种 | 森林种 | 挽用型 | 乘用型 | 兼用型 | 冷血统 | 热血统 | 温血统 | |
| 代表品种 | 蒙古马、哈萨克马 | 阿拉伯马、阿克哈-塔克马 | 百色马、宁强马、卡巴金马 | 雅库特马、爱沙尼亚马 | 阿尔登马、比 利时重挽马 | 纯血马、阿拉伯马 | 三河马 | 阿尔登马、苏维埃重挽马 | 阿拉伯马、阿克哈-塔克马 | 阿尔登马、苏维埃重挽马 |
| 特点 | 分布在世界上广阔的草原区,由于年昼夜栖息于草原上过群牧生活 | 生活在干燥沙漠地带的马,体格较轻 | 性格机敏,善走山路 | 生活在气候较冷的森林地带的马 | 挽用 | 乘用 | 未经过专门化选育的地方品种 | 类型为中悍或下悍,主要用于农用或运输 | 主要是指高度培育、较悍威、血统纯正的轻型马品种 | 体型高大,性情温顺,马术用马 |
表2
现代马主要品种介绍"
| | 品种 | |||||||
|---|---|---|---|---|---|---|---|---|
| 奎特马[ (Quarter horse) | 美国花马[66] (American Paint horse) | 安达卢西亚马[66] (Andalusian horse) | 田纳西走马[66] (Tennessee Walker horse) | 摩根马[66] (Morgan horse) | 汉诺威马[66] (Hanoverian horse) | 奥尔洛夫快步马[66] (Orlov Trotter horse) | ||
| 别称 | 四分之一英里马 | 油漆马 | 无 | 无 | 无 | 无 | 无 | |
| 体型 | 轻型马,150~160 cm | 轻型马,150~160 cm | 轻型马, 约152 cm | 轻型马,150~160 cm | 轻型马,142~152 cm | 轻型马, 约162 cm | 轻型马, 约160 cm | |
| 毛发 | 各种单一颜色 | 花色 | 骝色、灰色 | 各种单一颜色 | 单一颜色,灰色除外 | 各种单一颜色 | 灰色、黑色、骝色 | |
| 地理起源 | 美国 | 美国 | 西班牙 | 美国 | 美国 | 德国 | 俄罗斯 | |
| 特点 | 头部短而宽,口吻短,耳小,鼻孔大,眼睛间距宽,颌骨宽而界限分明。颈丰满、中等长度,颈脊薄。髋部较宽,臀腰部肌肉发达。 | 体型强壮有力,腰臀部强壮,体被大面积且不规则的斑块,每匹花马都具有独特的毛色。 | 颈部短,肌肉发达,肩部宽阔而健壮,速度不是很快,但机敏好动,波浪卷的鬃毛,世界上最古老也是最纯正的马种之一。 | 头干燥,耳朵小,颈长。肩和腰角长而斜,背短,肷强健,躯干比较短,给人一种方形的外貌。 | 尾长而优雅,耳小,性情温顺且稳定,外形精致紧凑,腰臀发育非常好,跗关节距离地面较低。 | 中等大小的头,胸部富有肌肉,胸线较深,有力且对称的四肢和大的关节,是有名的跳跃表演马和花式骑术表演马。 | 颈部长,肩隆较低背部直,身体长,臀腰部宽阔有力,四肢长,体型高而轻,整体的比例关系很协调。 | |
| 分布 | 美国 | 美国 | 西班牙赫雷斯 | 美国田纳西州 | 美国麻萨诸塞州和佛蒙特州 | 德国汉诺威 | 北部欧亚大陆 | |
| 别称 | 蒙古野马 | 百色石山矮马 | 西汉,乌孙马[ | 无 | 无 | 无 | 无 | |
| 体型 | 126~134 cm | 小型马,<106 cm | 131~139 cm | 轻型马, 144~152 cm | 轻型马, 约152 cm | 轻型马, 160~162 cm | 重型马, 162~172 cm | |
| 毛发 | 青色、骝色、黑色较多 | 骝色较多,粟毛、青毛、黑毛等次之 | 骝色、栗色、黑色较多,青色次之 | 各种单一颜色 | 金属般的栗色 | 各种单一颜色 | 黑色、骝色、灰色 | |
| 地理 起源 | 中国 | 中国 | 中国 | 阿拉伯半岛 | 土库曼斯坦 | 英国 | 英国 | |
| 特点 | 可分为乌珠穆沁马、百岔马、乌审马、阿巴嘎马、锡尼河马等。头大颈短,前胸丰满、胸深肋拱圆,腹大,腰背平直,四肢粗短,身低躯广。 | 头长而清秀,额宽适中,眼大而圆,鼻翼开张灵活,颈长短适中,胸宽而深,腹部圆大,蹄质坚实。 | 骨骼粗实、结构匀称,头中等大、略长,显粗重,多直颈,腰背平直,腹部圆大,关节明显,后躯发育较差,肷稍长,是一种古老的地方品种。 | 头小清秀,眼大,颈直,额宽,肩隆不突起,略凹的背部,肚围较深,四肢细,对世界上其他家马的形成具有重要影响。 | 头细颈高,眼大,两耳间的距离很宽,长、窄、管状的身体,肌肉发达,腿细而长,能在沙漠中越过特别长的距离,是跳跃和花式骑术用马。 | 体质干燥细致,身悍力强,骨骼修长,身材结实,腰臀有力,后腿长,速度快,是非常优秀的骑乘马品种,最有商业价值的品种之一。 | 两眼间距较宽,眼睛大而和善,鼻子为凸型,颈部较长,肩部宽,身体短而厚实,以力量著称,可能是挽马品种中最重的一种。 | |
| 分布 | 中国内蒙古自治区,蒙古高原等地 | 中国广西壮族自治区 | 中国新疆维吾尔自治区 | 阿拉伯半岛,沙特阿拉伯,也门等地 | 北部欧亚大陆,土库曼斯坦 | 英国 | 英国英格兰,米德兰 | |
| [1] | Wang YX, eds. A Complete Checklist of Mammal Species and Subspecies in China— A Taxonomic and Geographic Reference. Beijing: China Forestry Publishing House, 2003. |
| 王应祥著. 中国哺乳动物种和亚种分类名录与分布大全. 北京: 中国林业出版社, 2003. | |
| [2] |
Outram AK, Stear NA, Bendrey R, Olsen S, Kasparov A, Zaibert V, Thorpe N, Evershed RP. The earliest horse harnessing and milking. Science, 2009, 323(5919):1332-1335.
doi: 10.1126/science.1168594 pmid: 19265018 |
| [3] |
Gaunitz C, Fages A, Hanghøj K, Albrechtsen A, Khan N, Schubert M, Seguin-Orlando A, Owens IJ, Felkel S, Bignon-Lau O, de Barros Damgaard P, Mittnik A, Mohaseb AF, Davoudi H, Alquraishi S, Alfarhan AH, Al-Rasheid KAS, Crubézy E, Benecke N, Olsen S, Brown D, Anthony D, Massy K, Pitulko V, Kasparov A, Brem G, Hofreiter M, Mukhtarova G, Baimukhanov N, Lõugas L, Onar V, Stockhammer PW, Krause J, Boldgiv B, Undrakhbold S, Erdenebaatar D, Lepetz S, Mashkour M, Ludwig A, Wallner B, Merz V, Merz I, Zaibert V, Willerslev E, Librado P, Outram AK, Orlando L. Ancient genomes revisit the ancestry of domestic and Przewalski's horses. Science, 2018, 360(6384):111-114.
doi: 10.1126/science.aao3297 |
| [4] |
Fages A, Hanghøj K, Khan N, Gaunitz C, Seguin-Orlando A, Leonardi M, McCrory Constantz C, Gamba C, Al-Rasheid KAS, Albizuri S, Alfarhan AH, Allentoft M, Alquraishi S, Anthony D, Baimukhanov N, Barrett JH, Bayarsaikhan J, Benecke N, Bernáldez-Sánchez E, Berrocal-Rangel L, Biglari F, Boessenkool S, Boldgiv B, Brem G, Brown D, Burger J, Crubézy E, Daugnora L, Davoudi H, de Barros Damgaard P, de Los Ángeles de Chorro Y de Villa-Ceballos M, Deschler-Erb S, Detry C, Dill N, do Mar Oom M, Dohr A, Ellingvåg S, Erdenebaatar D, Fathi H, Felkel S, Fernández-Rodríguez C, García-Viñas E, Germonpré M, Granado JD, Hallsson JH, Hemmer H, Hofreiter M, Kasparov A, Khasanov M, Khazaeli R, Kosintsev P, Kristiansen K, Kubatbek T, Kuderna L, Kuznetsov P, Laleh H, Leonard JA, Lhuillier J, von Lettow-Vorbeck CL, Logvin A, Lõugas L, Ludwig A, Luis C, Arruda AM, Marques-Bonet T, Matoso Silva R, Merz V, Mijiddorj E, Miller BK, Monchalov O, Mohaseb FA, Morales A, Nieto-Espinet A, Nistelberger H, Onar V, Pálsdóttir AH, Pitulko V, Pitskhelauri K, Pruvost M, Rajic Sikanjic P, Rapan Papeša A, Roslyakova N, Sardari A, Sauer E, Schafberg R, Scheu A, Schibler J, Schlumbaum A, Serrand N, Serres-Armero A, Shapiro B, Sheikhi Seno S, Shevnina I, Shidrang S, Southon J, Star B, Sykes N, Taheri K, Taylor W, Teegen WR, Trbojević Vukičević T, Trixl S, Tumen D, Undrakhbold S, Usmanova E, Vahdati A, Valenzuela-Lamas S, Viegas C, Wallner B, Weinstock J, Zaibert V, Clavel B, Lepetz S, Mashkour M, Helgason A, Stefánsson K, Barrey E, Willerslev E, Outram AK, Librado P, Orlando L. Tracking five millennia of horse management with extensive ancient genome time series. Cell, 2019, 177(6): 1419-1435.e31.
doi: 10.1016/j.cell.2019.03.049 |
| [5] |
Orlando L, Ginolhac A, Raghavan M, Vilstrup J, Rasmussen M, Magnussen K, Steinmann KE, Kapranov P, Thompson JF, Zazula G, Froese D, Moltke I, Shapiro B, Hofreiter M, Al-Rasheid KAS, Gilbert MTP, Willerslev E. True single-molecule DNA sequencing of a pleistocene horse bone. Genome Res, 2011, 21(10):1705-1719.
doi: 10.1101/gr.122747.111 |
| [6] |
Orlando L, Ginolhac A, Zhang GJ, Froese D, Albrechtsen A, Stiller M, Schubert M, Cappellini E, Petersen B, Moltke I, Johnson PLF, Fumagalli M, Vilstrup JT, Raghavan M, Korneliussen T, Malaspinas AS, Vogt J, Szklarczyk D, Kelstrup CD, Vinther J, Dolocan A, Stenderup J, Velazquez AMV, Cahill J, Rasmussen M, Wang XL, Min JM, Zazula GD, Seguin-Orlando A, Mortensen C, Magnussen K, Thompson JF, Weinstock J, Gregersen K, Røed KH, Eisenmann V, Rubin CJ, Miller DC, Antczak DF, Bertelsen MF, Brunak S, Al-Rasheid KAS, Ryder O, Andersson L, Mundy J, Krogh A, Gilbert MTP, Kjær K, Sicheritz-Ponten T, Jensen LJ, Olsen JV, Hofreiter M, Nielsen R, Shapiro B, Wang J, Willerslev E. Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature, 2013, 499(7456):74-78.
doi: 10.1038/nature12323 |
| [7] | Schubert M, Jónsson H, Chang D, Der Sarkissian C, Ermini L, Ginolhac A, Albrechtsen A, Dupanloup I, Foucal A, Petersen B, Fumagalli M, Raghavan M, Seguin-Orlando A, Korneliussen TS, Velazquez AMV, Stenderup J, Hoover CA, Rubin CJ, Alfarhan AH, Alquraishi SA, Al-Rasheid KAS, MacHugh DE, Kalbfleisch T, MacLeod JN, Rubin EM, Sicheritz-Ponten T, Andersson L, Hofreiter M, Marques-Bonet T, Gilbert MTP, Nielsen R, Excoffier L, Willerslev E, Shapiro B, Orlando L. Prehistoric genomes reveal the genetic foundation and cost of horse domestication. Proc Natl Acad Sci USA, 2014, 111(52):E5661-5669. |
| [8] |
Cantalapiedra JL, Prado JL, Fernández MH, Alberdi MT. Decoupled ecomorphological evolution and diversification in Neogene-Quaternary horses. Science, 2017, 355(6325):627-630.
doi: 10.1126/science.aag1772 pmid: 28183978 |
| [9] |
Aberle KS, Hamann H, Drögemüller C, Distl O. Genetic diversity in German draught horse breeds compared with a group of primitive, riding and wild horses by means of microsatellite DNA markers. Anim Genet, 2004, 35(4):270-277.
pmid: 15265065 |
| [10] |
Aranguren-Méndez J, Jordana J, Gomez M. Genetic diversity in Spanish donkey breeds using microsatellite DNA markers. Genet Sel Evol, 2001, 33(4):433-442.
pmid: 11559485 |
| [11] |
Kusliy MA, Vorobieva NV, Tishkin AA, Makunin AI, Druzhkova AS, Trifonov VA, Iderkhangai T-O, Graphodatsky AS. Traces of Late Bronze and Early Iron Age Mongolian horse mitochondrial lineages in modern populations. Genes (Basel), 2021, 12(3):412.
doi: 10.3390/genes12030412 |
| [12] |
Wallner B, Palmieri N, Vogl C, Rigler D, Bozlak E, Druml T, Jagannathan V, Leeb T, Fries R, Tetens J, Thaller G, Metzger J, Distl O, Lindgren G, Rubin CJ, Andersson L, Schaefer R, McCue M, Neuditschko M, Rieder S, Schlötterer C, Brem G. Y chromosome uncovers the recent oriental origin of modern stallions. Curr Biol, 2017, 27(13): 2029-2035.e5.
doi: 10.1016/j.cub.2017.05.086 |
| [13] |
Chopineau M, Stewart F, Allen WR. Cloning and analysis of the cDNA encoding the horse and donkey luteinizing hormone beta-subunits. Gene, 1995, 160(2):253-256.
pmid: 7642105 |
| [14] |
Tallmadge RL, Lear TL, Johnson AK, Guérin G, Millon LV, Carpenter SL, Antczak DF. Characterization of the beta2-microglobulin gene of the horse. Immunogenetics, 2003, 54(10):725-733.
pmid: 12557059 |
| [15] |
Wade CM, Giulotto E, Sigurdsson S, Zoli M, Gnerre S, Imsland F, Lear TL, Adelson DL, Bailey E, Bellone RR, Blöcker H, Distl O, Edgar RC, Garber M, Leeb T, Mauceli E, MacLeod JN, Penedo MCT, Raison JM, Sharpe T, Vogel J, Andersson L, Antczak DF, Biagi T, Binns MM, Chowdhary BP, Coleman SJ, Della Valle G, Fryc S, Guérin G, Hasegawa T, Hill EW, Jurka J, Kiialainen A, Lindgren G, Liu J, Magnani E, Mickelson JR, Murray J, Nergadze SG, Onofrio R, Pedroni S, Piras MF, Raudsepp T, Rocchi M, Røed KH, Ryder OA, Searle S, Skow L, Swinburne JE, Syvänen AC, Tozaki T, Valberg SJ, Vaudin M, White JR, Zody MC, Broad Institute Genome Sequencing Platform, Broad Institute Whole Genome Assembly Team, Lander ES, Lindblad-Toh K,. Genome sequence, comparative analysis, and population genetics of the domestic horse. Science, 2009, 326(5954):865-867.
doi: 10.1126/science.1178158 pmid: 19892987 |
| [16] |
McCue ME, Bannasch DL, Petersen JL, Gurr J, Bailey E, Binns MM, Distl O, Guérin G, Hasegawa T, Hill EW, Leeb T, Lindgren G, Penedo MCT, Røed KH, Ryder OA, Swinburne JE, Tozaki T, Valberg SJ, Vaudin M, Lindblad-Toh K, Wade CM, Mickelson JR. A high density SNP array for the domestic horse and extant Perissodactyla: utility for association mapping, genetic diversity, and phylogeny studies. PLoS Genet, 2012, 8(1):e1002451.
doi: 10.1371/journal.pgen.1002451 |
| [17] |
Schaefer RJ, Schubert M, Bailey E, Bannasch DL, Barrey E, Bar-Gal GK, Brem G, Brooks SA, Distl O, Fries R, Finno CJ, Gerber V, Haase B, Jagannathan V, Kalbfleisch T, Leeb T, Lindgren G, Lopes MS, Mach N, da Câmara Machado A, MacLeod JN, McCoy A, Metzger J, Penedo C, Polani S, Rieder S, Tammen I, Tetens J, Thaller G, Verini-Supplizi A, Wade CM, Wallner B, Orlando L, Mickelson JR, McCue ME. Developing a 670k genotyping array to tag ~2M SNPs across 24 horse breeds. BMC Genomics, 2017, 18(1):565.
doi: 10.1186/s12864-017-3943-8 pmid: 28750625 |
| [18] |
Doan R, Cohen ND, Sawyer J, Ghaffari N, Johnson CD, Dindot SV. Whole-genome sequencing and genetic variant analysis of a Quarter horse mare. BMC Genomics, 2012, 13:78.
doi: 10.1186/1471-2164-13-78 |
| [19] |
Warmuth V, Eriksson A, Bower MA, Barker G, Barrett E, Hanks BK, Li SC, Lomitashvili D, Ochir-Goryaeva M, Sizonov GV, Soyonov V, Manica A. Reconstructing the origin and spread of horse domestication in the Eurasian steppe. Proc Natl Acad Sci USA, 2012, 109(21):8202-8206.
doi: 10.1073/pnas.1111122109 |
| [20] |
Nergadze SG, Lupotto M, Pellanda P, Santagostino M, Vitelli V, Giulotto E. Mitochondrial DNA insertions in the nuclear horse genome. Anim Genet, 2010, 41 Suppl 2: 176-185.
doi: 10.1111/age.2010.41.issue-s2 |
| [21] |
Jansen T, Forster P, Levine MA, Oelke H, Hurles M, Renfrew C, Weber J, Olek K. Mitochondrial DNA and the origins of the domestic horse. Proc Natl Acad Sci USA, 2002, 99(16):10905-10910.
doi: 10.1073/pnas.152330099 |
| [22] |
Achilli A, Olivieri A, Soares P, Lancioni H, Hooshiar Kashani B, Perego UA, Nergadze SG, Carossa V, Santagostino M, Capomaccio S, Felicetti M, Al-Achkar W, Penedo MCT, Verini-Supplizi A, Houshmand M, Woodward SR, Semino O, Silvestrelli M, Giulotto E, Pereira L, Bandelt HJ, Torroni A. Mitochondrial genomes from modern horses reveal the major haplogroups that underwent domestication. Proc Natl Acad Sci USA, 2012, 109(7):2449-2454.
doi: 10.1073/pnas.1111637109 |
| [23] |
McGahern A, Bower MAM, Edwards CJ, Brophy PO, Sulimova G, Zakharov I, Vizuete-Forster M, Levine M, Li S, MacHugh DE, Hill EW. Evidence for biogeographic patterning of mitochondrial DNA sequences in Eastern horse populations. Anim Genet, 2006, 37(5):494-497.
pmid: 16978180 |
| [24] |
Lei CZ, Su R, Bower MA, Edwards CJ, Wang XB, Weining S, Liu L, Xie WM, Li F, Liu RY, Zhang YS, Zhang CM, Chen H. Multiple maternal origins of native modern and ancient horse populations in China. Anim Genet, 2009, 40(6):933-944.
doi: 10.1111/j.1365-2052.2009.01950.x pmid: 19744143 |
| [25] |
Wallner B, Piumi F, Brem G, Müller M, Achmann R. Isolation of Y chromosome-specific microsatellites in the horse and cross-species amplification in the genus Equus. J Hered, 2004, 95(2):158-164.
pmid: 15073232 |
| [26] |
Wallner B, Vogl C, Shukla P, Burgstaller JP, Druml T, Brem G. Identification of genetic variation on the horse y chromosome and the tracing of male founder lineages in modern breeds. PLoS One, 2013, 8(4):e60015.
doi: 10.1371/journal.pone.0060015 |
| [27] |
Ling YH, Ma YH, Guan WJ, Cheng YJ, Wang YP, Han JL, Jin DP, Mang L, Mahmut H. Identification of Y chromosome genetic variations in Chinese indigenous horse breeds. J Hered, 2010, 101(5):639-643.
doi: 10.1093/jhered/esq047 |
| [28] |
Kakoi H, Kikuchi M, Tozaki T, Hirota KI, Nagata SI, Hobo S, Takasu M. Distribution of Y chromosomal haplotypes in Japanese native horse populations. J Equine Sci, 2018, 29(2):39-42.
doi: 10.1294/jes.29.39 |
| [29] |
Librado P, Khan N, Fages A, Kusliy MA, Suchan T, Tonasso-Calvière L, Schiavinato S, Alioglu D, Fromentier A, Perdereau A, Aury JM, Gaunitz C, Chauvey L, Seguin-Orlando A, Der Sarkissian C, Southon J, Shapiro B, Tishkin AA, Kovalev AA, Alquraishi S, Alfarhan AH, Al-Rasheid KAS, Seregély T, Klassen L, Iversen R, Bignon-Lau O, Bodu P, Olive M, Castel JC, Boudadi- Maligne M, Alvarez N, Germonpré M, Moskal-Del Hoyo M, Wilczyński J, Pospuła S, Lasota-Kuś A, Tunia K, Nowak M, Rannamäe E, Saarma U, Boeskorov G, Lōugas L, Kyselý R, Peške L, Bălășescu A, Dumitrașcu V, Dobrescu R, Gerber D, Kiss V, Szécsényi-Nagy A, Mende BG, Gallina Z, Somogyi K, Kulcsár G, Gál E, Bendrey R, Allentoft ME, Sirbu G, Dergachev V, Shephard H, Tomadini N, Grouard S, Kasparov A, Basilyan AE, Anisimov MA, Nikolskiy PA, Pavlova EY, Pitulko V, Brem G, Wallner B, Schwall C, Keller M, Kitagawa K, Bessudnov AN, Bessudnov A, Taylor W, Magail J, Gantulga JO, Bayarsaikhan J, Erdenebaatar D, Tabaldiev K, Mijiddorj E, Boldgiv B, Tsagaan T, Pruvost M, Olsen S, Makarewicz CA, Lamas SV, Canadell SA, Espinet AN, Iborra MP, Garrido JL, González ER, Celestino S, Olària C, Arsuaga JL, Kotova N, Pryor A, Crabtree P, Zhumatayev R, Toleubaev A, Morgunova NL, Kuznetsova T, Lordkipanize D, Marzullo M, Prato O, Gianni GB, Tecchiati U, Clavel B, Lepetz S, Davoudi H, Mashkour M, Berezina NY, Stockhammer PW, Krause J, Haak W, Morales-Muñiz A, Benecke N, Hofreiter M, Ludwig A, Graphodatsky AS, Peters J, Kiryushin KY, Iderkhangai TO, Bokovenko NA, Vasiliev SK, Seregin NN, Chugunov KV, Plasteeva NA, Baryshnikov GF, Petrova E, Sablin M, Ananyevskaya E, Logvin A, Shevnina I, Logvin V, Kalieva S, Loman V, Kukushkin I, Merz I, Merz V, Sakenov S, Varfolomeyev V, Usmanova E, Zaibert V, Arbuckle B, Belinskiy AB, Kalmykov A, Reinhold S, Hansen S, Yudin AI, Vybornov AA, Epimakhov A, Berezina NS, Roslyakova N, Kosintsev PA, Kuznetsov PF, Anthony D, Kroonen GJ, Kristiansen K, Wincker P, Outram A, Orlando L. The origins and spread of domestic horses from the Western Eurasian steppes. Nature, 2021, 598(7882):634-640.
doi: 10.1038/s41586-021-04018-9 |
| [30] |
Kader A, Li Y, Dong KZ, Irwin DM, Zhao QJ, He XH, Liu JF, Pu YB, Gorkhali NA, Liu XX, Jiang L, Li XC, Guan WJ, Zhang YP, Wu DD, Ma YH. Population variation reveals independent selection toward small body size in Chinese Debao pony. Genome Biol Evol, 2015, 8(1):42-50.
doi: 10.1093/gbe/evv245 |
| [31] |
Petersen JL, Mickelson JR, Rendahl AK, Valberg SJ, Andersson LS, Axelsson J, Bailey E, Bannasch D, Binns MM, Borges AS, Brama P, da Câmara Machado A, Capomaccio S, Cappelli K, Cothran EG, Distl O, Fox-Clipsham L, Graves KT, Guérin G, Haase B, Hasegawa T, Hemmann K, Hill EW, Leeb T, Lindgren G, Lohi H, Lopes MS, McGivney BA, Mikko S, Orr N, Penedo MCT, Piercy RJ, Raekallio M, Rieder S, Røed KH, Swinburne J, Tozaki T, Vaudin M, Wade CM, McCue ME. Genome-wide analysis reveals selection for important traits in domestic horse breeds. PLoS Genet, 2013, 9(1):e1003211.
doi: 10.1371/journal.pgen.1003211 |
| [32] |
Petersen JL, Mickelson JR, Cothran EG, Andersson LS, Axelsson J, Bailey E, Bannasch D, Binns MM, Borges AS, Brama P, da Câmara Machado A, Distl O, Felicetti M, Fox-Clipsham L, Graves KT, Guérin G, Haase B, Hasegawa T, Hemmann K, Hill EW, Leeb T, Lindgren G, Lohi H, Lopes MS, McGivney BA, Mikko S, Orr N, Penedo MCT, Piercy RJ, Raekallio M, Rieder S, Røed KH, Silvestrelli M, Swinburne J, Tozaki T, Vaudin M, Wade CM, McCue ME. Genetic diversity in the modern horse illustrated from genome-wide SNP data. PLoS One, 2013, 8(1):e54997.
doi: 10.1371/journal.pone.0054997 |
| [33] |
Ling YH, Ma YH, Guan WJ, Cheng YJ, Wang YP, Han JL, Mang L, Zhao QJ, He XH, Pu YB, Fu BL. Evaluation of the genetic diversity and population structure of Chinese indigenous horse breeds using 27 microsatellite markers. Anim Genet, 2011, 42(1):56-65.
doi: 10.1111/j.1365-2052.2010.02067.x pmid: 20477800 |
| [34] | China National Commission of Resources, eds. Animal Genetic Resources in China. Horses, Donkeys and Camels. Beijing: China Agriculture Press, 2011. |
| 国家畜禽遗传资源委员会组编. 中国畜禽遗传资源志·马驴驼志. 北京: 中国农业出版社, 2011. | |
| [35] |
Lippold S, Matzke NJ, Reissmann M, Hofreiter M. Whole mitochondrial genome sequencing of domestic horses reveals incorporation of extensive wild horse diversity during domestication. BMC Evol Biol, 2011, 11:328.
doi: 10.1186/1471-2148-11-328 |
| [36] |
Librado P, Gamba C, Gaunitz C, Der Sarkissian C, Pruvost M, Albrechtsen A, Fages A, Khan N, Schubert M, Jagannathan V, Serres-Armero A, Kuderna LFK, Povolotskaya IS, Seguin-Orlando A, Lepetz S, Neuditschko M, Thèves C, Alquraishi S, Alfarhan AH, Al-Rasheid K, Rieder S, Samashev Z, Francfort HP, Benecke N, Hofreiter M, Ludwig A, Keyser C, Marques- Bonet T, Ludes B, Crubézy E, Leeb T, Willerslev E, Orlando L. Ancient genomic changes associated with domestication of the horse. Science, 2017, 356(6336):442-445.
doi: 10.1126/science.aam5298 pmid: 28450643 |
| [37] |
Der Sarkissian C, Ermini L, Schubert M, Yang MA, Librado P, Fumagalli M, Jónsson H, Bar-Gal GK, Albrechtsen A, Vieira FG, Petersen B, Ginolhac A, Seguin-Orlando A, Magnussen K, Fages A, Gamba C, Lorente-Galdos B, Polani S, Steiner C, Neuditschko M, Jagannathan V, Feh C, Greenblatt CL, Ludwig A, Abramson NI, Zimmermann W, Schafberg R, Tikhonov A, Sicheritz-Ponten T, Willerslev E, Marques-Bonet T, Ryder OA, McCue M, Rieder S, Leeb T, Slatkin M, Orlando L. Evolutionary genomics and conservation of the endangered Przewalski's horse. Curr Biol, 2015, 25(19):2577-2583.
doi: 10.1016/j.cub.2015.08.032 pmid: 26412128 |
| [38] |
Cruz F, Vilà C, Webster MT. The legacy of domestication: accumulation of deleterious mutations in the dog genome. Mol Biol Evol, 2008, 25(11):2331-2336.
doi: 10.1093/molbev/msn177 |
| [39] |
Lu J, Tang T, Tang H, Huang JZ, Shi SH, Wu CI. The accumulation of deleterious mutations in rice genomes: a hypothesis on the cost of domestication. Trends Genet, 2006, 22(3):126-131.
doi: 10.1016/j.tig.2006.01.004 |
| [40] |
Lin T, Zhu GT, Zhang JH, Xu XY, Yu QH, Zheng Z, Zhang ZH, Lun YY, Li S, Wang XX, Huang ZJ, Li JM, Zhang CZ, Wang TT, Zhang YY, Wang AX, Zhang YC, Lin K, Li CY, Xiong GS, Xue YB, Mazzucato A, Causse M, Fei ZJ, Giovannoni JJ, Chetelat RT, Zamir D, Städler T, Li JF, Ye ZB, Du YC, Huang SW. Genomic analyses provide insights into the history of tomato breeding. Nat Genet, 2014, 46(11):1220-1226.
doi: 10.1038/ng.3117 |
| [41] |
McCue ME, Valberg SJ, Lucio M, Mickelson JR. Glycogen synthase 1 (GYS1) mutation in diverse breeds with polysaccharide storage myopathy. J Vet Intern Med, 2008, 22(5):1228-1233.
doi: 10.1111/j.1939-1676.2008.0167.x pmid: 18691366 |
| [42] | Bowling AT. Horse Genetics. Oxfordshire: Cab International, 1996. |
| [43] |
Rees JL. Genetics of hair and skin color. Annu Rev Genet, 2003, 37:67-90.
doi: 10.1146/genet.2003.37.issue-1 |
| [44] |
Furumura M, Sakai C, Abdel-Malek Z, Barsh GS, Hearing VJ. The interaction of agouti signal protein and melanocyte stimulating hormone to regulate melanin formation in mammals. Pigment Cell Res, 1996, 9(4):191-203.
doi: 10.1111/pcr.1996.9.issue-4 |
| [45] |
Rieder S, Taourit S, Mariat D, Langlois B, Guérin G. Mutations in the agouti (ASIP), the extension (MC1R), and the brown (TYRP1) loci and their association to coat color phenotypes in horses (Equus caballus). Mamm Genome, 2001, 12(6):450-455.
doi: 10.1007/s003350020017 pmid: 11353392 |
| [46] | Bailey E, Brooks SA. Chapter4: Black, Bay and Chestnut (Extension and Agouti). In: Horse Genetics. Boston, CABI. 2013. |
| [47] |
Mariat D, Taourit S, Guérin G. A mutation in the MATP gene causes the cream coat colour in the horse. Genet Sel Evol, 2003, 35(1):119-133.
pmid: 12605854 |
| [48] |
Cook D, Brooks S, Bellone R, Bailey E. Missense mutation in exon 2 of SLC36A1 responsible for champagne dilution in horses. PLoS Genet, 2008, 4(9):e1000195.
doi: 10.1371/journal.pgen.1000195 |
| [49] |
Brunberg E, Andersson L, Cothran G, Sandberg K, Mikko S, Lindgren G. A missense mutation in PMEL17 is associated with the silver coat color in the horse. BMC Genet, 2006, 7:46.
pmid: 17029645 |
| [50] |
Imsland F, McGowan K, Rubin CJ, Henegar C, Sundström E, Berglund J, Schwochow D, Gustafson U, Imsland P, Lindblad-Toh K, Lindgren G, Mikko S, Millon L, Wade C, Schubert M, Orlando L, Penedo MCT, Barsh GS, Andersson L. Regulatory mutations in TBX3 disrupt asymmetric hair pigmentation that underlies Dun camouflage color in horses. Nat Genet, 2016, 48(2):152-158.
doi: 10.1038/ng.3475 pmid: 26691985 |
| [51] | Zhao RY, Zhao YP, Li B, Bou G, Zhang XZ, Mongke T, Bao T, Gereliin S, Gereltuuin T, Li C, Bai DY, Dugarjaviin M. Overview of the genetic control of horse coat color patterns. Hereditas(Beijing), 2018, 40(5):357-368. |
| 赵若阳, 赵一萍, 李蓓, 格日乐其木格, 张心壮, 陶克涛, 图格琴, 旭仁其木格, 青柏, 李超, 白东义, 芒来. 马毛色遗传机理研究进展. 遗传, 2018, 40(5):357-368. | |
| [52] |
Promerová M, Andersson LS, Juras R, Penedo MCT, Reissmann M, Tozaki T, Bellone R, Dunner S, Hořín P, Imsland F, Imsland P, Mikko S, Modrý D, Roed KH, Schwochow D, Vega-Pla JL, Mehrabani-Yeganeh H, Yousefi-Mashouf N, Cothran EG, Lindgren G, Andersson L. Worldwide frequency distribution of the 'Gait keeper' mutation in the DMRT3 gene. Anim Genet, 2014, 45(2):274-282.
doi: 10.1111/age.12120 pmid: 24444049 |
| [53] |
Andersson LS, Larhammar M, Memic F, Wootz H, Schwochow D, Rubin CJ, Patra K, Arnason T, Wellbring L, Hjälm G, Imsland F, Petersen JL, McCue ME, Mickelson JR, Cothran G, Ahituv N, Roepstorff L, Mikko S, Vallstedt A, Lindgren G, Andersson L, Kullander K. Mutations in DMRT3 affect locomotion in horses and spinal circuit function in mice. Nature, 2012, 488(7413):642-646.
doi: 10.1038/nature11399 |
| [54] |
Hill EW, Gu J, McGivney BA, MacHugh DE. Targets of selection in the Thoroughbred genome contain exercise- relevant gene SNPs associated with elite racecourse performance. Anim Genet, 2010, 41 Suppl 2: 56-63.
doi: 10.1111/age.2010.41.issue-s2 |
| [55] |
Hill EW, Gu JJ, Eivers SS, Fonseca RG, McGivney BA, Govindarajan P, Orr N, Katz LM, MacHugh DE. A sequence polymorphism in MSTN predicts sprinting ability and racing stamina in thoroughbred horses. PLoS One, 2010, 5(1):e8645.
doi: 10.1371/journal.pone.0008645 |
| [56] |
Schröder W, Klostermann A, Stock KF, Distl O. A genome-wide association study for quantitative trait loci of show-jumping in Hanoverian warmblood horses. Anim Genet, 2012, 43(4):392-400.
doi: 10.1111/j.1365-2052.2011.02265.x pmid: 22497689 |
| [57] |
Liu XX, Zhang YL, Li YF, Pan JF, Wang DD, Chen WH, Zheng ZQ, He XH, Zhao QJ, Pu YB, Guan WJ, Han JL, Orlando L, Ma YH, Jiang L. EPAS1 gain-of-function mutation contributes to high-altitude adaptation in Tibetan horses. Mol Biol Evol, 2019, 36(11):2591-2603.
doi: 10.1093/molbev/msz158 |
| [58] |
Makvandi-Nejad S, Hoffman GE, Allen JJ, Chu E, Gu E, Chandler AM, Loredo AI, Bellone RR, Mezey JG, Brooks SA, Sutter NB. Four loci explain 83% of size variation in the horse. PLoS One, 2012, 7(7):e39929.
doi: 10.1371/journal.pone.0039929 |
| [59] |
Orr N, Back W, Gu J, Leegwater P, Govindarajan P, Conroy J, Ducro B, Van Arendonk JAM, MacHugh DE, Ennis S, Hill EW, Brama PAJ. Genome-wide SNP association-based localization of a dwarfism gene in Friesian dwarf horses. Anim Genet, 2010, 41 Suppl 2: 2-7.
doi: 10.1111/age.2010.41.issue-s2 |
| [60] |
Nanaei HA, Esmailizadeh A, Mehrgardi AA, Han JL, Wu DD, Li Y, Zhang YP. Comparative population genomic analysis uncovers novel genomic footprints and genes associated with small body size in Chinese pony. BMC Genomics, 2020, 21(1):496.
doi: 10.1186/s12864-020-06887-2 |
| [61] |
Sutter NB, Bustamante CD, Chase K, Gray MM, Zhao KY, Zhu L, Padhukasahasram B, Karlins E, Davis S, Jones PG, Quignon P, Johnson GS, Parker HG, Fretwell N, Mosher DS, Lawler DF, Satyaraj E, Nordborg M, Lark KG, Wayne RK, Ostrander EA. A single IGF1 allele is a major determinant of small size in dogs. Science, 2007, 316(5821):112-115.
doi: 10.1126/science.1137045 |
| [62] |
Lango AH, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, Willer CJ, Jackson AU, Vedantam S, Raychaudhuri S, Ferreira T, Wood AR, Weyant RJ, Segrè AV, Speliotes EK, Wheeler E, Soranzo N, Park JH, Yang J, Gudbjartsson D, Heard-Costa NL, Randall JC, Qi L, Vernon Smith A, Mägi R, Pastinen T, Liang LM, Heid IM, Luan JA, Thorleifsson G, Winkler TW, Goddard ME, Sin Lo K, Palmer C, Workalemahu T, Aulchenko YS, Johansson A, Zillikens MC, Feitosa MF, Esko T, Johnson T, Ketkar S, Kraft P, Mangino M, Prokopenko I, Absher D, Albrecht E, Ernst F, Glazer NL, Hayward C, Hottenga JJ, Jacobs KB, Knowles JW, Kutalik Z, Monda KL, Polasek O, Preuss M, Rayner NW, Robertson NR, Steinthorsdottir V, Tyrer JP, Voight BF, Wiklund F, Xu JF, Zhao JH, Nyholt DR, Pellikka N, Perola M, Perry JRB, Surakka I, Tammesoo ML, Altmaier EL, Amin N, Aspelund T, Bhangale T, Boucher G, Chasman DI, Chen C, Coin L, Cooper MN, Dixon AL, Gibson Q, Grundberg E, Hao K, Juhani Junttila M, Kaplan LM, Kettunen J, König IR, Kwan T, Lawrence RW, Levinson DF, Lorentzon M, McKnight B, Morris AP, Müller M, Suh Ngwa J, Purcell S, Rafelt S, Salem RM, Salvi E, Sanna S, Shi JX, Sovio U, Thompson JR, Turchin MC, Vandenput L, Verlaan DJ, Vitart V, White CC, Ziegler A, Almgren P, Balmforth AJ, Campbell H, Citterio L, De Grandi A, Dominiczak A, Duan JB, Elliott P, Elosua R, Eriksson JG, Freimer NB, Geus EJC, Glorioso N, Shen HQ, Hartikainen AL, Havulinna AS, Hicks AA, Hui J, Igl W, Illig T, Jula A, Kajantie E, Kilpeläinen TO, Koiranen M, Kolcic I, Koskinen S, Kovacs P, Laitinen J, Liu JJ, Lokki ML, Marusic A, Maschio A, Meitinger T, Mulas A, Paré G, Parker AN, Peden JF, Petersmann A, Pichler I, Pietiläinen KHP, Pouta A, Ridderstråle M, Rotter JI, Sambrook JG, Sanders AR, Schmidt CO, Sinisalo J, Smit JH, Stringham HM, Bragi Walters G, Widen E, Wild SH, Willemsen G, Zagato L, Zgaga L, Zitting P, Alavere H, Farrall M, McArdle WL, Nelis M, Peters MJ, Ripatti S, van Meurs JBJ, Aben KK, Ardlie KG, Beckmann JS, Beilby JP, Bergman RN, Bergmann S, Collins FS, Cusi D, den Heijer M, Eiriksdottir G, Gejman PV, Hall AS, Hamsten A, Huikuri HV, Iribarren C, Kähönen M, Kaprio J, Kathiresan S, Kiemeney L, Kocher T, Launer LJ, Lehtimäki T, Melander O, Mosley TH, Musk AW, Nieminen MS, O'Donnell CJ, Ohlsson C, Oostra B, Palmer LJ, Raitakari O, Ridker PM, Rioux JD, Rissanen A, Rivolta C, Schunkert H, Shuldiner AR, Siscovick DS, Stumvoll M, Tönjes A, Tuomilehto J, van Ommen GJ, Viikari J, Heath AC, Martin NG, Montgomery GW, Province MA, Kayser M, Arnold AM, Atwood LD, Boerwinkle E, Chanock SJ, Deloukas P, Gieger C, Grönberg H, Hall P, Hattersley AT, Hengstenberg C, Hoffman W, Lathrop GM, Salomaa V, Schreiber S, Uda M, Waterworth D, Wright AF, Assimes TL, Barroso I, Hofman A, Mohlke KL, Boomsma DI, Caulfield MJ, Cupples LA, Erdmann J, Fox CS, Gudnason V, Gyllensten U, Harris TB, Hayes RB, Jarvelin MR, Mooser V, Munroe PB, Ouwehand WH, Penninx BW, Pramstaller PP, Quertermous T, Rudan I, Samani NJ, Spector TD, Völzke H, Watkins H, Wilson JF, Groop LC, Haritunians T, Hu FB, Kaplan RC, Metspalu A, North KE, Schlessinger D, Wareham NJ, Hunter DJ, O'Connell JR, Strachan DP, Wichmann HE, Borecki IB, van Duijn CM, Schadt EE, Thorsteinsdottir U, Peltonen L, Uitterlinden AG, Visscher PM, Chatterjee N, Loos RJF, Boehnke M, McCarthy MI, Ingelsson E, Lindgren CM, Abecasis GR, Stefansson K, Frayling TM, Hirschhorn JN. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature, 2010, 467(7317):832-838.
doi: 10.1038/nature09410 |
| [63] |
Boyko AR, Quignon P, Li L, Schoenebeck JJ, Degenhardt JD, Lohmueller KE, Zhao KY, Brisbin A, Parker HG, vonHoldt BM, Cargill M, Auton A, Reynolds A, Elkahloun AG, Castelhano M, Mosher DS, Sutter NB, Johnson GS, Novembre J, Hubisz MJ, Siepel A, Wayne RK, Bustamante CD, Ostrander EA. A simple genetic architecture underlies morphological variation in dogs. PLoS Biol, 2010, 8(8):e1000451.
doi: 10.1371/journal.pbio.1000451 |
| [64] |
Flori L, Fritz S, Jaffrézic F, Boussaha M, Gut I, Heath S, Foulley JL, Gautier M. The genome response to artificial selection: a case study in dairy cattle. PLoS One, 2009, 4(8):e6595.
doi: 10.1371/journal.pone.0006595 |
| [65] | Gansu Agricultural University, eds . Horse Raising. Beijing: Agriculture Press, 1981. |
| 甘肃农业大学主编. 养马学. 北京: 农业出版社, 1981. | |
| [66] | (English) Edwards EH. Eyewitness handbooks-horse. Beijing: Chinese Friendship Press Co., 1998. |
| (英)爱德华兹. 猫头鹰出版社译. 全世界100多种马匹的彩色图鉴. 北京: 中国友谊出版公司, 1998. |
| [1] | 刘羽诚, 申妍婷, 田志喜. 大豆泛基因组研究进展[J]. 遗传, 2024, 46(3): 183-198. |
| [2] | 赵洪, 薛勇彪. 显花植物自交不亲和性的分子与演化机制[J]. 遗传, 2024, 46(1): 3-17. |
| [3] | 田璐妍, 黄小珍. 植物开花调控中蛋白质相分离机制在从头驯化中的应用价值[J]. 遗传, 2023, 45(9): 754-764. |
| [4] | 廉小平, 黄光福, 张玉娇, 张静, 胡凤益, 张石来. 长雄野生稻有利基因的发掘与利用[J]. 遗传, 2023, 45(9): 765-780. |
| [5] | 简六梅, 肖英杰, 严建兵. 从头驯化:作物品种设计与培育的新方向[J]. 遗传, 2023, 45(9): 741-753. |
| [6] | 赖笔威, 陈磊, 芦思佳. 大豆光周期适应性研究进展[J]. 遗传, 2023, 45(9): 793-800. |
| [7] | 邢超凡, 王闽涛, 王磊, 申欣. 两侧对称动物左右不对称发生机制研究进展[J]. 遗传, 2023, 45(6): 488-500. |
| [8] | 姜明亮, 郎红, 李晓楠, 祖野, 赵靖, 彭沈凌, 刘振, 战宗祥, 朴钟云. 植物孤基因研究进展[J]. 遗传, 2022, 44(8): 682-694. |
| [9] | 文子龙, 赵毅强. 群体遗传学下动物驯化研究进展[J]. 遗传, 2021, 43(3): 226-239. |
| [10] | 高志伟, 王龙. 真核生物起源研究进展[J]. 遗传, 2020, 42(10): 929-948. |
| [11] | 杨新萍,于媛,许操. 重新设计与快速驯化创造新型作物[J]. 遗传, 2019, 41(9): 827-835. |
| [12] | 林春,刘正杰,董玉梅,MichelVales,毛自朝. 藜麦的驯化栽培与遗传育种[J]. 遗传, 2019, 41(11): 1009-1022. |
| [13] | 赵永欣, 李孟华, 赵要风. 中国绵羊起源、进化和遗传多样性研究进展[J]. 遗传, 2017, 39(11): 958-973. |
| [14] | 潘章源, 贺小云, 王翔宇, 郭晓飞, 曹晓涵, 胡文萍, 狄冉, 刘秋月, 储明星. 家养动物选择信号研究进展[J]. 遗传, 2016, 38(12): 1069-1080. |
| [15] | 张焕萍, 尹佟明. 谱系特有基因研究进展[J]. 遗传, 2015, 37(6): 544-553. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||

www.chinagene.cn
备案号:京ICP备09063187号-4
总访问:,今日访问:,当前在线:

